The Arms Race between Transposable Elements and KRAB-ZFPs and its Impact on Mammals

- Todd S. Macfarlan, PhD, Head, Section on Mammalian Development and Evolution
- Sherry Ralls, BA, Biologist
- Melania Bruno, PhD, Visiting Fellow
- Jinpu Jin, PhD, Visiting Fellow
- Mohamed Mahgoub, PhD, Visiting Fellow
- Anna Dorothea Senft, PhD, Visiting Fellow
- Ming-an Sun, PhD, Visiting Fellow
- Michael B. Evans, DO, Clinical Fellow
- Steven Gay, MD, Clinical Fellow
- Anne Martini, DO, Clinical Fellow
- Justin Demmerle, BA, Graduate Student
- Amanda Ivanoff, BS, Postbaccalaureate Fellow
- Catrina Rateb, BS, BA, Postbaccalaureate Fellow
At the NICHD, our central mission is to ensure that every human is born healthy. Despite much progress in understanding the many ways the mother interacts with the fetus during development, we still know little about the molecular changes that promoted the emergence of placental mammals from our egg-laying relatives over 100 million years ago, nor about those mechanisms that continue to drive phenotypic differences amongst mammals. One attractive hypothesis is that retroviruses and their endogenization into the genomes of our ancestors played an important role in eutherian evolution, by providing protein-coding genes such as syncytins (derived from retroviral env genes that cause cell fusions in placental trophoblasts) and novel gene-regulatory nodes that altered expression networks, to allow for implantation and the emergence and continued evolution of the placenta. Our primary interest is to explore the impact of such endogenous retroviruses (ERVs), which account for about 10% of our genomic DNA, on embryonic development and on the evolution of new traits in mammals. This has led us to examine the rapidly evolving Kruppel-associated box zinc-finger protein (KZFP) family, the single largest family of transcription factors (TFs) in most, if not all, mammalian genomes. Our hypothesis is that KZFP gene expansion and diversification was driven primarily by the constant onslaught of ERVs and other transposable elements (TEs) to the genomes of our ancestors, as a means to transcriptionally repress them. The hypothesis is supported by recent evidence demonstrating that the majority of KZFPs bind to TEs and that TEs and nearby genes are activated in KZFP knockout mice. We will continue to explore the impacts of the TE/KZFP “arms race” on the evolution of mammals. We will also begin a new phase of our research to explore whether KZFPs play broader roles in genome regulation, beyond gene silencing, and how such functions impact mammalian development and evolution.
Kruppel-associated box zinc-finger (ZF) proteins (KRAB-ZFPs) have emerged as candidates that recognize ERVs. KRAB-ZFPs are rapidly evolving transcriptional repressors that emerged in a common ancestor of coelacanths, birds, and tetrapods, and they constitute the largest family of transcription factors in mammals (estimated to be several hundred in mice and humans). Each species has its own unique repertoire of KRAB-ZFPs, with a small number shared by closely related species and a larger fraction specific to each species. Despite their abundance, little is known about their physiological functions. KRAB-ZFPs consist of an N-terminal KRAB domain that binds to the co-repressor KAP1 and a variable number of C-terminal C2H2 ZF domains that mediate sequence-specific DNA binding. KAP1 directly interacts with the KRAB domain, which recruits the histone methyltransferase (HMT) SETDB1 and heterochromatin protein 1 (HP1) to initiate heterochromatic silencing. Several lines of evidence point to a role for the KRAB-ZFP family in ERV silencing. First, the number of C2H2 ZF genes in mammals correlates with the number of ERVs. Second, the KRAB-ZFP protein ZFP809 was isolated based on its ability to bind to the primer binding site for proline tRNA (PBSpro) of murine leukemia virus (MuLV). Third, deletion of the KRAB-ZFP co-repressors Trim28 or Setdb1 leads to activation of many ERVs. We have therefore begun a systematic interrogation of KRAB-ZFP function as a potential adaptive repression system against ERVs.
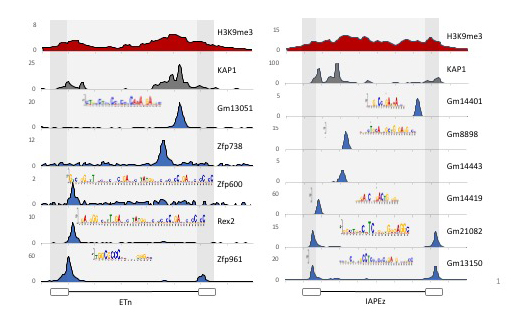
Figure 1. KRAB-ZFPs bind redundantly to the active retrotransposons ETn and IAP.
ChIP-seq track for indicated KRAB-ZFPs displayed across a consensus sequence of ETn and IAP retrotransposons. ChIP-seq was performed by expressing epitope-tagged KRAB-ZFPs in F9 ECs or ESCs and ChIP'ing with antibodies that recognize the epitope.
We initially focused on ZFP809 as a likely ERV–suppressing KRAB-ZFP, given that it was originally identified as part of a repression complex that recognizes infectious MuLV via direct binding to the 18 nt Primer Binding Site for Proline (PBSpro) sequence. We hypothesized that ZFP809 functions in vivo to repress other ERVs that utilize the PBSpro. Using ChIP-seq of epitope-tagged ZFP809 in embryonic stems cells (ESCs) and embryonic carcinoma (EC) cells, we determined that ZFP809 binds to several sub-classes of ERV elements via the PBSpro. We generated Zfp809 knockout mice to determine whether ZFP809 was required for silencing the ERV element VL30pro. We found that Zfp809 knockout tissues displayed high levels of VL30pro elements and that the targeted elements display an epigenetic shift from repressive epigenetic marks (H3K9me3 and CpG methylation) to active marks (H3K9Ac and CpG hypo-methylation). ZFP809–mediated repression extended to a handful of genes that contained adjacent VL30pro integrations. Furthermore, using a combination of conditional alleles and rescue experiments, we determined that ZFP809 activity was required in development to initiate silencing, but not in somatic cells to maintain silencing. The studies provided the first demonstration of the in vivo requirement of a KRAB-ZFP in the recognition and silencing of ERVs.
As a follow-up to our studies on ZFP809, we began a systematic analysis of KRAB-ZFPs using a medium-throughput ChIP-seq screen and functional genomics of KRAB-ZFP clusters and individual KRAB-ZFP genes. Our ChIP-seq data demonstrate that the majority of recently evolved KRAB-ZFP genes interact with and repress distinct and partially overlapping ERVs and other retrotransposons targets. The hypothesis is strongly supported by the distinct ERV reactivation phenotypes we observed in mouse ESC lines lacking one of five of the largest KRAB-ZFP gene clusters. Furthermore, our preliminary evidence suggests that KRAB-ZFP cluster knockout (KO) mice are viable, but have elevated rates of somatic retrotransposition of specific retrotransposon families, providing the first direct genetic link between KRAB-ZFP gene diversification and retrotransposon mobility.
Although our data show that many KRAB-ZFPs repress ERVs, we also found that more ancient KRAB-ZFPs, which emerged in a human/mouse common ancestor, do not bind to or repress ERVs. One such KRAB-ZFP, ZFP568, plays an important role in silencing a key developmental gene that may have played a critical role in the onset of viviparity in mammals. Using ChIP-seq and biochemical assays, we determined that ZFP568 is a direct repressor of a placental-specific isoform of the Igf2 gene called Igf2-P0. Insulin-like growth factor 2 (Igf2) is the major fetal growth hormone in mammals. We demonstrated that loss of Zfp568, which causes gastrulation failure, or mutation of the ZFP568 binding site at the Igf2-P0 promoter, cause inappropriate Igf2-P0 activation. We also showed that the lethality could be rescued by deletion of Igf2. The data highlight the exquisite selectivity by which members of the KRAB-ZFP family repress their targets, and they identify an additional layer of transcriptional control of a key growth factor regulating fetal and placental development. In a follow-up to these studies, we determined that ZFP568 is highly conserved and under purifying selection in eutheria with the exception of human. Human ZNF568 allele variants have lost the ability to bind to and repress Igf2-P0, which may have been driven by the loss of the Igf2-p0 transcript in human placenta. We solved the crystal structure of mouse ZFP568 ZFs bound to the Igf2-P0 binding site, which reveals several non-canonical ZF–DNA contacts, highlighting the ability of individual ZFs to change confirmation depending upon ZF context and DNA structure. The structures also explain how mutations in human ZNF568 alleles disrupt Igf2-P0 interactions, which contain either deleted ZFs or mutations of key ZF-DNA contact residues. Taken together, our studies provide important insights into the evolutionary and structural dynamics of ZF-DNA interactions, which play a key role in regulating mammalian development and evolution.
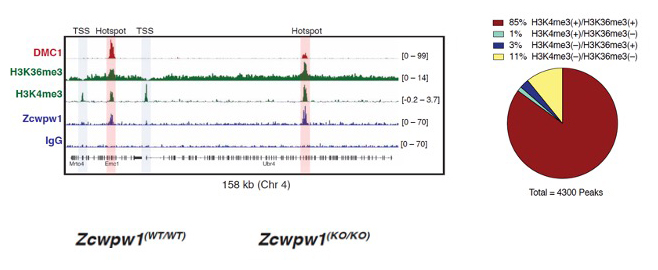
Figure 2. ZCWPW1 binds to meiotic recombination hotspots in spermatocytes harboring dual H3K4me3 and H3K36me3 marks.
ChIP-seq or Cut & Run was performed with indicated antibodies in mouse spermatocytes, and read pileup is displayed across a region on Chr 4. The pie chart at the right displays percentage of ZCWPW1 peaks that overlap peaks of either H3K4me3, H3K36me3, or both marks.
We began a new exploration of the function and mechanism of PRDM9, the most ancient KRAB-ZFP, which emerged in the jawless fish and which plays a highly specialized role in meiotic recombination (MR). MR generates genetic diversity in sexually reproducing organisms and ensures proper synapsis and segregation of homologous chromosomes in gametes. Errors in MR that lead to mis-segregation of chromosomes are a leading cause of miscarriage and childhood disease. MR is initiated by programmed double-strand breaks (DSBs) in DNA that are distributed non-randomly at thousands of specific 1–2 kb regions called hotspots. In most mammals, hotspots are defined by PRDM9, a protein that contains a rapidly evolving DNA–binding ZF array and a specialized histone methyltransferase activity that catalyzes dual trimethylation marks on histone H3 at lysine 4 and 36 (H3K4me3 and H3K36me3), whose activities are both required for hotspot specification. Prdm9 loss-of-function causes sterility in mice, and PRDM9 mutations have been associated with male infertility in humans. In species lacking Prdm9, including yeast, plants, and birds, hotspots are located in H3K4me3–rich regions at gene promoters. Thus, the emergence of PRDM9 during evolution reshaped the MR landscape by relocating DSBs away from promoters to chromatin sites bound by the rapidly evolving PRDM9, which allowed for rapid interspecies hotspot diversification. We set out to address whether other factors, in addition to PRDM9, are required to “re-engineer” hotspot selection and how the DNA break and repair machinery is recruited to sites marked by PRDM9. We identified the dual histone methylation readers Zcwpw1, which co-evolved with and is tightly co-expressed with Prdm9. Using a mouse model, we found that ZCWPW1 is an essential meiotic recombination factor required for efficient repair of PRDM9–dependent DSBs and pairing homologous chromosomes in males mice. However, ZCWPW1 is not required for the initiation of DSBs at PRDM9 binding sites. Our results indicate that the evolution of a dual histone methylation writer (PRDM9) and reader (ZCWPW1) system in vertebrates remodeled genetic recombination hotspot selection from an ancestral static pattern near genes towards a flexible pattern controlled by the rapidly evolving DNA–binding activity of PRDM9.
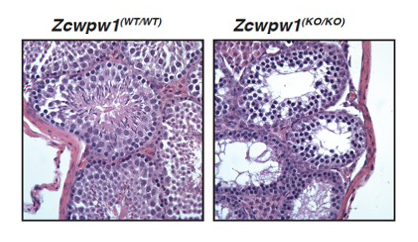
Figure 3. Zcwpw1 KO mice are azoospermic.
H&E staining of adult testes from WT or Zcwpw1 KO mice. Sperm are completely lacking in Zcwpw1 mutants due to defective meiotic double-strand-break repair.
Additional Funding
- NICHD Strategic Planning Award: The role of THAP7 in neurodevelopment and intellectual disability
Publications
- Mahgoub M, Paiano J, Bruno M, Wu W, Pathuri S, Zhang X, Ralls S, Cheng X, Nussenzweig A,Macfarlan TS. Dual histone methyl reader ZCWPW1 facilitates repair of meiotic double strand breaks in male mice. eLife 2020;9:e53360.
- Wolf G, de Iaco A, Sun MA, Bruno M, Tinkham M, Hoang D, Mitra A, Ralls S, Trono D, Macfarlan TS. KRAB-zinc finger protein gene expansion in response to active retrotransposons in the murine lineage. eLife 2020;9:e56337.
- Sun M, Wolf G, Wang Y, Ralls S, Senft AD, Jin J, Dunn-Fletcher CE, Muglia LJ, Macfarlan TS. Deciphering the evolution of the transcriptional and regulatory landscape in human placenta. BioRxiv 2020; https://doi.org/10.1101/2020.09.11.289686.
- Haase AD, Macfarlan TS. PiRNAs rise to rescue koalas. Cell 2019;179(3):582-583.
- Chen C, Sun MA, Warzecha C, Bachu M, Dey A, Wu T, Adams PD, Macfarlan T, Love P, Ozato K. HIRA, a DiGeorge syndrome candidate gene, confers proper chromatin accessibility on HSCs and supports all stages of hematopoiesis. Cell Rep 2020;30(7):2136-2149.
Collaborators
- Xiaodong Cheng, PhD, Emory University, Atlanta, GA
Contact
For more information, email todd.macfarlan@nih.gov or visit http://macfarlan.nichd.nih.gov.