Quantitative Imaging and Tissue Sciences

- Peter J. Basser, PhD, Head, Section on Quantitative Imaging and Tissue Sciences
- Ferenc Horkay, PhD, Staff Scientist
- Julian Rey, PhD, Postdoctoral Intramural Research Training Award Fellow
- Nathan Williamson, PhD, Postdoctoral Research Associate Training Award Fellow (NIGMS)
- Velencia Witherspoon, PhD, Postdoctoral Intramural Research Training Award Fellow
- Teddy Cai, BS, Predoctoral Intramural Research Training Award Fellow, NIH OxCam Program
- Kristofor Pas, BS, Postbaccalaureate Intramural Research Training Award Fellow
- Alexandru Avram, PhD, Collaborating Scientist funded via the Henry Jackson Foundation and the Center for Neuroscience and Regenerative Medicine
- Dan Benjamini, PhD, Collaborating Scientist funded via the Henry Jackson Foundation and the Center for Neuroscience and Regenerative Medicine
- Michal Komlosh, PhD, Collaborating Scientist funded via the Henry Jackson Foundation and the Center for Neuroscience and Regenerative Medicine
- Magdoom Kulam, PhD, Collaborating Scientist funded via the Henry Jackson Foundation and the Center for Neuroscience and Regenerative Medicine
- Kadharbatcha Saleem, PhD, Collaborating Scientist funded via the Henry Jackson Foundation and the Center for Neuroscience and Regenerative Medicine
- Alexandros Chremos, PhD, Contract Scientist
- Rea Ravin, PhD, Contract Scientist
In our tissue-sciences research, we strive to understand fundamental relationships between function and structure in living tissues. Specifically, we are interested in how tissue microstructure, hierarchical organization, composition, and material properties all work together to affect biological function or dysfunction. We investigate biological and physical model systems at various length and time scales, performing biophysical measurements and developing novel physical/mathematical models (including molecular dynamics [MD] and continuum models) to explain their functional properties and behavior. Inextricably connected to these activities is our study of water and its interactions with macromolecules and ions in biological media. Experimentally, we often use water to probe tissue structure and function from nanometers to centimeters and from microseconds to lifetimes. Our armamentarium includes atomic force microscopy (AFM), small-angle X-ray scattering (SAXS), small-angle neutron scattering (SANS), static light scattering (SLS), dynamic light scattering (DLS), osmometry, and multi-dimensional nuclear magnetic resonance (NMR) relaxometry and diffusometry. A goal is to develop understanding and tools that can be translated from bench-based quantitative methodologies to the bedside to aid in diagnosis and therapy.
Our activities dovetail with our basic and applied research in quantitative imaging, which is intended to generate measurements and maps of intrinsic physical quantities, including diffusivities, relaxivities, exchange rates, etc., rather than the qualitative ‘weighted’ MR images conventionally used in radiology. At a basic level, our work is directed toward making critical ‘invisible’ biological structures and processes ‘visible.’ Our quantitative imaging group uses knowledge of physics, engineering, applied mathematics, imaging and computer sciences, as well as key insights gleaned from our tissue-sciences research, to discover, vet, and develop novel quantitative imaging biomarkers that can detect changes in tissue composition, microstructure, and/or microdynamics with high sensitivity and specificity. The ultimate translational goal is to assess normal and abnormal developmental trajectories, diagnose childhood diseases and disorders, and characterize degeneration and trauma (such as mild traumatic brain injury). MRI is our imaging modality of choice because it is so well suited to many applications critical to the NICHD mission; it is non-invasive, non-ionizing, usually requires no exogenous contrast agents or dyes, and is generally deemed safe and effective for use with mothers, fetuses, and children in both clinical and research settings. Critical to this enterprise is our ability to follow water as it diffuses through complex media as a probe of microstructure, and to assess its interactions with biomolecules to identify distinct water compartments in tissues.
One of our translational goals has been to transform clinical MRI scanners into scientific instruments capable of producing reproducible, accurate, and precise imaging data with which to measure and map useful imaging biomarkers for various clinical applications, including single scans, longitudinal, and multi-site studies, personalized medicine, and genotype/phenotype correlation studies, as well as for populating imaging databases with high-quality normative data. From a more basic perspective, another goal has been to apply our various MRI tools and methodologies to advance neuroscience, providing new methods to explore brain structure/function relationships, such as “imaging” the human connectome.
Figure 1. Digital “D99” Macaque Brain Atlas
Kadharbatcha Saleem created this NHP (non-human primate) brain atlas, which identifies deep gray matter areas, such as the thalamus, using both histological staining and Mean Apparent Propagator MR–derived imaging parameters. Careful correlation and integration of histology and MR images is critical to testing and vetting the sensitivity and specificity of new MR brain imaging methods. In some cases, MRI methods can be more eloquent in identifying nuclei and other structures than staining.
In vivo MRI histology
The most mature in vivo MRI histological technology that we invented, developed, and clinically translated is Diffusion Tensor MRI (DTI), by which we measure and map D, a diffusion tensor of water, within an imaging volume. Information derived from this quantity includes white-matter fiber-tract orientation, the orientationally averaged mean apparent diffusion constant (mADC) or mean diffusivity (MD), and other intrinsic scalar (invariant) quantities. Such imaging parameters have been used by radiologists and neuroscientists as non-invasive quantitative histological ‘stains’ that are obtained by probing endogenous tissue water in vivo without requiring any exogenous contrast agents or dyes. The mADC is the most widely used diffusion imaging parameter in neuroradiology to identify ischemic areas in the brain during acute stroke and to follow cancer patients’ responses to therapy. The measures of diffusion anisotropy we first proposed (e.g., the fractional anisotropy or FA) are also widely used to follow changes in normally and abnormally developing white matter and in many other clinical and neuroscience applications, such as brain white-matter visualization. Our group also pioneered the use of fiber direction–encoded color (DEC) maps to display the orientation of white matter pathways in the brain. To assess anatomical connectivity among various cortical and deep-brain gray-matter areas, we also proposed and developed DTI ‘Streamline’ Tractography, which is used to track white-matter fibers to help establish ‘anatomical connectivity’ and by neuroradiologists and neurosurgeons, so as to plan surgical procedures, and radiation dosing, so as to spare ‘eloquent’ areas of the brain. These advances also helped inspire several large federally funded research initiatives, including the NIH Human Connectome Project (HCP) and, more recently, the NIH Brain Initiative.
More recently, we invented and developed a family of advanced in vivo diffusion MRI methods to measure fine-scale microstructural features of axons and fascicles (also known as ‘microstructure imaging’), which otherwise could only be assessed using laborious ex vivo histological or pathological methods. We have been developing efficient means for performing ‘k- and q-space MRI’ in the living human brain, such as ‘Mean Apparent Propagator’ (MAP) MRI, an approach that can detect subtle microstructural and architectural features in both gray and white matter at micron-scale resolution, several orders of magnitude smaller than the typical MRI voxel. MAP MRI also subsumes DTI, as well as providing a bevy of new in vivo quantitative imaging biomarkers to measure and map. We recently applied this to assessing mild traumatic brain injury (TBI) and other forms of TBI. We also developed CHARMED MRI, which measures the average axon diameter (AAD), and AxCaliber MRI, which measures the axon-diameter distribution (ADD) along white-matter pathways, and we reported the first in vivo measurement of ADDs within the rodent corpus callosum. The ADD is functionally important, given that axon diameter is a critical determinant of conduction velocity and therefore the rate at which information is transferred along axon bundles, and helps determine the latencies or time delays between and among different brain areas. This led us to propose a novel MRI–based method to measure the ‘latency connectome,’ including a latency matrix that reports conduction times between different brain areas. We also developed a companion mathematical theory to explain the observed ADDs in different fascicles, suggesting that they represent a trade-off between maximizing information flow and minimizing metabolic demands. We developed novel multiple pulsed-field gradient (mPFG) methods and demonstrated their feasibility in vivo on conventional clinical MRI scanners as a further means to extract quantitative features in the central nervous system (CNS), such as the AAD and other features of cell size and shape.
Figure 2. Elucidating cortical layers using MAP MRI–derived parameters
Cortical layers are generally difficult to identify using conventional MRI methods, but MAP MRI provides a family of stains that can be used to identify microstructural differences among cortical layers, allowing for non-invasive parcellation of some brain areas. The image shows maps of the diffusion-encoded color (DEC), Fractional Anisotropy (FA), Mean Diffusivity (MD), Non-gaussianity (NG), Propagator Anisotropy (PA), and Return to Axis Probability (RTAP). Shown also are the cortical layers numbered, using the convention of 1 through 6 from the cortex to the sub-cortical white matter.
Although gray matter appears featureless in the brain with DTI, its microstructure and architecture are rich and varied throughout the brain, not only along the brain's cortical surface, but also within and among its various cortical layers and within deep gray-matter regions. To target this tissue, we have been developing several non-invasive, in vivo methods to measure unique features of cortical gray-matter microstructure and architecture that are visible in electron microscopy (EM) applications but currently invisible in conventional MRI. One example is diffusion tensor distribution (DTD) MRI, in which we use our normal tensor-variate distribution to characterize heterogeneities in these complex tissues. One of our long-term goals is to ‘parcellate’ or segment the cerebral cortex in vivo into its approximately 500 distinct cyto-architechtonic areas, using non-invasive imaging methods. To this end, we are developing advanced MRI sequences and analysis pipelines to probe correlations among relaxivities and diffusivities of different water pools in the cortex. One promising avenue is to use multi-dimensional MRI relaxometry– and diffusometry–based methods to study water mobility and diffusion in gray and white matter. We continue to work to translate these and other methods to the clinic to help identify changes in normal and abnormal development, as well as in inflammation and trauma. Along these lines, we made excellent progress this past year in developing radiological-pathological correlations between MR and neuropathological images of TBI tissue specimens as a way to identify potential quantitative imaging biomarkers of injury or inflammation that may have the potential to detect TBI in vivo.
Figure 3. Identifying boundaries between brain areas using micro-structural imaging methods
Mean Apparent Propagator (MAP) MRI–derived biomarkers are juxtaposed to images of the same specimen after being histologically stained in area F4. MR parameters are often able to detect boundaries between different cortical brain areas more reliably than by histology.
Quantitative MRI biomarker development for pediatric and fetal imaging applications
MRI is considered safer than X-ray–based methods, such as computed tomography (CT), for scanning fetuses, infants, and children. However, clinical MRI still lacks the quantitative character of CT. The scope of conventional MRI clinical applications is limited to revealing either gross morphological features or focal abnormalities. Clinical MRI also often lacks the biological specificity necessary for developing robust and reliable imaging ‘biomarkers.’ In particular, MRI assessment of normal brain development and developmental disorders has benefited greatly from the introduction of ‘quantitative’ clinical MRI techniques, with which one measures and maps meaningful intrinsic physical quantities or chemical variables that possess physical units and can be compared among different tissue regions. Quantitative MRI methods such as DTI also increase sensitivity, providing a basis for monitoring subtle changes that occur, e.g., during the progression or remission of disease, by comparing measurements in a single subject against normative values obtained from a healthy population. Quantitative MRI methods should continue to advance ‘precision imaging’ studies, in which MRI phenotypic and genotypic data can be meaningfully incorporated and used for improved diagnosis and prognosis assessments.
To advance our quantitative imaging activities, we developed numerical and statistical methods, including algorithms that generate a continuous, smooth approximation to the discrete, noisy, measured DTI field data, so as to reduce noise, and which allowed us to implement Streamline Tractography. We proposed a novel Gaussian distribution for the tensor-valued random variables that we use to design optimal DTI experiments and interpret their results. In tandem, we developed non-parametric empirical (e.g., Bootstrap) methods to determine the statistical distribution of DTI–derived quantities in order to study, e.g., the inherent variability and reliability of computed white-matter trajectories, enabling us to apply powerful hypothesis tests to assess the statistical significance of findings in a wide range of important biological and clinical applications that had been tested using ad hoc statistical methods. We are also developing novel methods to register different brain volumes and to generate group-average DTI data or atlases from various subject populations, based on the Kullback-Leibler divergence.
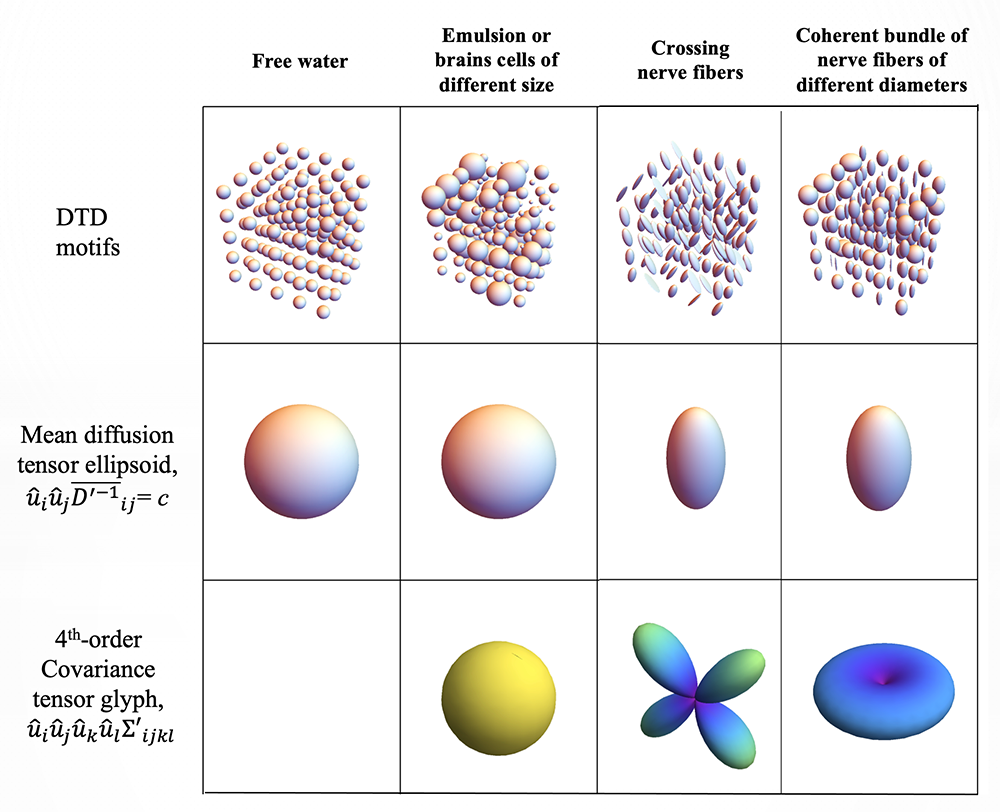
Figure 4. Identifying boundaries between brain areas using micro-structural imaging methods
While Diffusion Tensor MRI (DTI) provides an estimate of a mean diffusion tensor within a voxel, Diffusion Tensor Distribution (DTD) MRI reveals how heterogeneous the population of microdiffusion tensors is within the same voxel, providing a wealth of new information about local microstructural variability. For instance, different microtensor motifs can lead to the same voxel averaged mean diffusion tensor. The glyph of the covariance tensor captures this variability, making it possible to distinguish among these different motifs.
Previously, we carried out clinical studies that utilize novel quantitative MRI acquisition and analysis methods and whose aim is to improve accuracy and reproducibility of diagnosis and to detect and follow normal and abnormal development. One early example is the NIH Study of Normal Brain Development, jointly sponsored by the NICHD, NIMH, NINDS, and NIDA, which was initiated in 1998 and intended to advance our understanding of normal brain development in typical healthy children and adolescents. The Brain Development Cooperative Group is still actively publishing papers, primarily by mining the rich high-quality MRI data, many of which our lab processed, serving as the DTI Data-Processing Center (DPC). The processed DTI data collected from the project were uploaded into a database and made publicly available through the National Database for Autism Research (NDAR). Our collaborator Carlo Pierpaoli, who spearheaded this work, continues to support, update, and disseminate the processing and analysis software called “TORTOISE,” which grew out of this effort and which can be downloaded from http://www.tortoisedti.org.
Traumatic Brain Injury (TBI) represents a significant public health challenge for our pediatric population, but also for young men and women who serve in the military. Our involvement in TBI research, particularly in detecting mild TBI (mTBI), has continued to expand through partnerships with various Department of Defense (DoD) entities. Diffusion MRI (dMRI) provides essential information to aid in the assessment of TBI, but conventional dMRI methods have lacked sufficient specificity. To improve the accuracy and reproducibility of MAP–MRI findings, we developed a data–processing pipeline, and, in collaboration with scientists at the DoD Center for Neuroscience and Regenerative Medicine (CNRM), performed the first normative MAP–MRI studies, and applied this new and powerful method to detect tissue damage in brains of individuals who have suffered TBI, extending our NICHD TORTOISE pipeline to be able to analyze MAP–MRI data. We are now employing promising multi-dimensional MRI relaxometry-diffusometry methods to study the etiology of various types of TBI, in collaboration with the USUHS Neuropathology Research Division and under the auspices of the CNRM, and to improve the correlation and integration of neuropathology and neuroradiological imaging data, so as to speed the deployment of new MRI methods to assess TBI. We also partnered with CNRM to study ways to measure very slow flows that occur during glymphatic transport, a mechanism the brain uses to wash away harmful macromolecules, just as the lymphatic system uses in other organs. With our partners at the University of Arizona, this research is providing experimental data to enable us migrate these imaging approaches to the clinic, to be able to assess normal and pathological glymphatic transport in vivo.
We are also collaborating with Sara Inati, who studies focal epilepsy, a devastating disorder that is difficult to detect using conventional neuroradiological methods. We are developing and testing various new MRI–based methods that we believe may reveal pathological microstructural features and changes in architectural organization of the brain in this disorder, for example, in cortical dysplasia, to improve localization and assessment of cortical lesions.
We have been partnering with Roberto Romero and Mark Haacke to develop novel fetal brain MR imaging applications. Currently, it is challenging to measure quantitative imaging biomarkers in utero, particularly diffusion MRI–based ones, owing to large-scale fetal and maternal motion during the scanning session, and owing to the difficulty in acquiring image volumes with sufficient coverage, quality, and spatial resolution in a clinically feasible amount of time. Our lab has been developing novel approaches to address each of these critical issues. The suspension of clinical protocols owing to COVID-19 has reduced our data collection activities since March, 2020.
Biopolymer physics: water-ion-biopolymer interactions
Remarkably, despite their crucial role, little is known about the physical underpinnings of water-ion-biopolymer interactions, particularly in the physiological ionic strength regime. To determine the effect of ions on the structure and dynamics of key biopolymers, we developed a multi-scale experimental framework by combining macroscopic techniques (osmotic swelling-pressure measurements and mechanical measurements) with high-resolution scattering methods (e.g., SANS and SAXS), which probe the structure and interactions over a broad range of length and time scales. Macroscopic swelling-pressure measurements provide information on the overall thermodynamic response of the system, while SANS and SAXS allow us to investigate biopolymers at molecular and supramolecular length scales and to quantify the effect of changes in the environment (e.g., ion concentration, ion valance, pH, temperature) on the structure and interactions among biopolymers, water, and ions. Studies carried out on well defined model systems that mimic essential features of tissues provide important insights that cannot be obtained from experimental studies made on biological systems themselves. Mathematical models based on well established polymer-physics concepts and, more recently, molecular dynamics (MD) simulation approaches make it possible to design experiments to help us quantify and explain aspects of tissue behavior and thus the underlying molecular and macroscopic mechanisms that govern key aspects of a tissue's normal functional properties.
These basic studies have led to numerous novel MRI phantom designs to support our quantitative imaging program, including diffusion MRI phantoms, which we use to calibrate scanners to assure the quality and fidelity of the imaging data in single-subject, longitudinal and multi-site studies. For instance, our U.S. Patent for a ‘Phantom for diffusion MRI imaging’ is now being used in the CaliberMRI phantom, enabling quantitative diffusion MRI studies to be performed at a myriad of sites. Our colleagues at NIST Boulder have incorporated our polyvinylpyrrolidone (PVP) polymer into their own diffusion MRI NIST standard. We used various glass microcapillary geometries to mimic some features of white matter pathways and to interrogate our AxCaliber and dPFG MRI models. We also developed a variety of NMR and MRI phantoms, such as a 3-D printed polymer phantom, which possess various salient features of cell or tissue systems, such as microscopic anisotropy, providing data with which to test the validity of our models and experimental designs. We are also developing novel polymer gel phantoms to calibrate exchange experiments in which to follow water moving between different microenvironments.
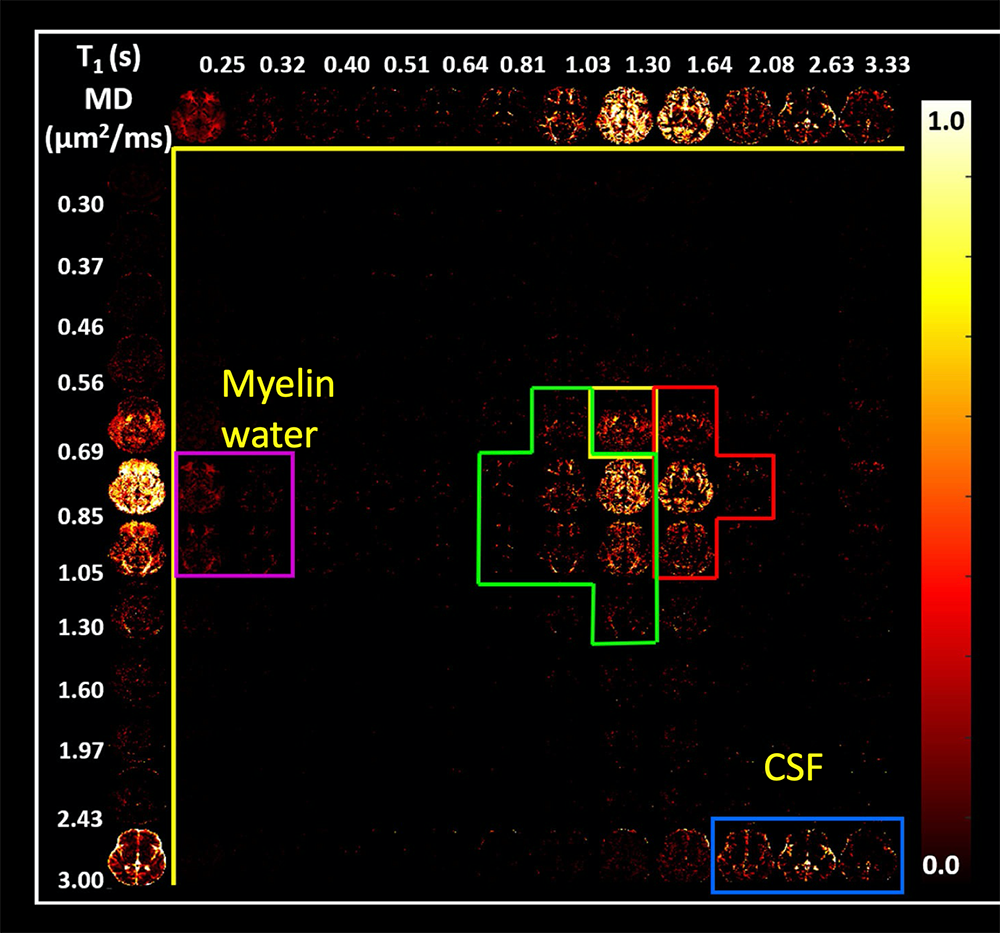
Figure 5. T1-MD MRI provides a powerful probe of different water environments within each voxel.
Images showing the correlation between relaxivity T1 and the mean diffusivity (MD) help identify distinct water domains or populations within voxels in the brain in which water may be in different chemical and physical environments. Such correlation spectra help identify white matter, gray matter, and cerebral spinal fluid compartments, as well as intra-myelin water, which is often difficult to detect.
Measuring and mapping functional properties of extracellular matrix (ECM)
We study interactions among the main extracellular matrix (ECM) components, often using cartilage as a model system because it is aneural, avascular, and almost acellular. In cartilage ECM, collagen (type II) is organized into fiber bundles that form a network that entraps the major proteoglycan (PG), a bottlebrush-shaped aggrecan. The biomechanical behavior of cartilage and other ECMs reflects their molecular composition and microstructure, which change during development, disease, degeneration, and aging. To determine tissue structure/function relationships, we measure various physical/chemical properties of ECM tissues and tissue analogs at different length and time scales, using a variety of complementary static and dynamic experimental techniques, e.g., osmometry, SANS, SAXS, neutron spin-echo (NSE), SLS, DLS, and AFM. Understanding the physical and chemical mechanisms affecting cartilage swelling (hydration) is essential to predicting its load-bearing ability, which is mainly governed by osmotic and electrostatic forces. To quantify the effect of hydration on cartilage properties, we previously developed a novel tissue micro-osmometer to perform precise and rapid measurements on small tissue samples (less than 1 microgram) as a function of the equilibrium water activity (vapor pressure). We also make osmotic pressure measurements to determine how the individual components of cartilage ECM (e.g., aggrecan and collagen) contribute to the total load-bearing capacity of the tissue. We also demonstrated that aggrecan-hyaluronic aggregates self-assemble into microgels, contributing to improved dimensional stability of the tissue and its lubricating ability. We further found that aggrecan is highly insensitive to changes in the ionic environment, particularly to divalent cations like calcium, which is critical for maintaining the tissue's mechanical integrity and allowing aggrecan to serve as a calcium-ion reservoir in cartilage and bone.
To model cartilage ECM, we invented and developed a new biomimetic composite material consisting of polyacrylic acid (PAA) microgel particles dispersed and embedded within a polyvinyl alcohol (PVA) gel matrix. PAA mimics the proteoglycan (i.e., hyaluronic-aggrecan complexes), while PVA mimics the fibrous collagen network entrapping them. Remarkably, the PVA/PAA biomimetic model system reproduces not only the shape of the cartilage swelling pressure curves, but also the numerical stiffness values reported for healthy and osteoarthritic human cartilage samples. Studies on these model composite hydrogels should continue to yield invaluable insights into the effects of various macromolecular factors (matrix stiffness, swelling pressure, fixed-charge density, etc.) on the tissue's macroscopic mechanical/swelling properties, and ultimately its remarkable load-bearing and lubricating abilities, and their loss in various diseases and disorders, including osteoarthritis.
We are now attempting to translate our understanding of the structure-function relationships of ECM components to develop and design novel non-invasive MRI methods, with the aim of inferring ECM composition, patency, and functional properties in vivo. Our goal is to use MRI for early diagnosis of diseases of cartilage and other tissue and organs to follow normal and abnormal ECM development, which entails making components of ECM (e.g., collagen and PGs) that are ‘invisible’ to MR ‘visible’ so as to predict the functional properties of the composite tissue, such as its load-bearing ability. An obstacle is that protons bound to immobile species (e.g., collagen) are largely invisible with conventional MRI methods. However, magnetization exchange (MEX) MRI (as well as other related methods) make it possible to detect the bound protons indirectly by transferring their magnetization to the abundant free water protons surrounding them. It also enables us to estimate collagen content in tissue quantitatively. In previous pilot studies with Uzi Eliav (deceased) and Ed Mertz, we applied the new MEX MRI method to determine the concentration and distribution of the main macromolecular constituents in bovine femoral-head cartilage samples. The results were qualitatively consistent with those obtained by histological techniques, such as high-definition infrared (HDIRI) spectroscopic imaging. Our novel approach has the potential to map tissue structure and functional properties in vivo and non-invasively. We are now developing molecular dynamics (MD)–based models of cartilage and cartilage ECM analogs in order to interpret our experimental findings, develop and test novel hypotheses, and predict the behavior of our model system under different experimental conditions.
Recently, we have been employing several novel MR methodologies that use our one-sided NMR systems to study water relaxation, diffusion, and exchange processes in ECM as a means to characterize its functional properties. Most recently, Velencia Witherspoon has been using these approaches to study the organization and structure of fascia. Our specialized NMR profilers are ideally suited to these tasks, as they can probe layered media, such as cartilage and fascia, using ultra-thin slices, almost as thin as an optical microscope provides.
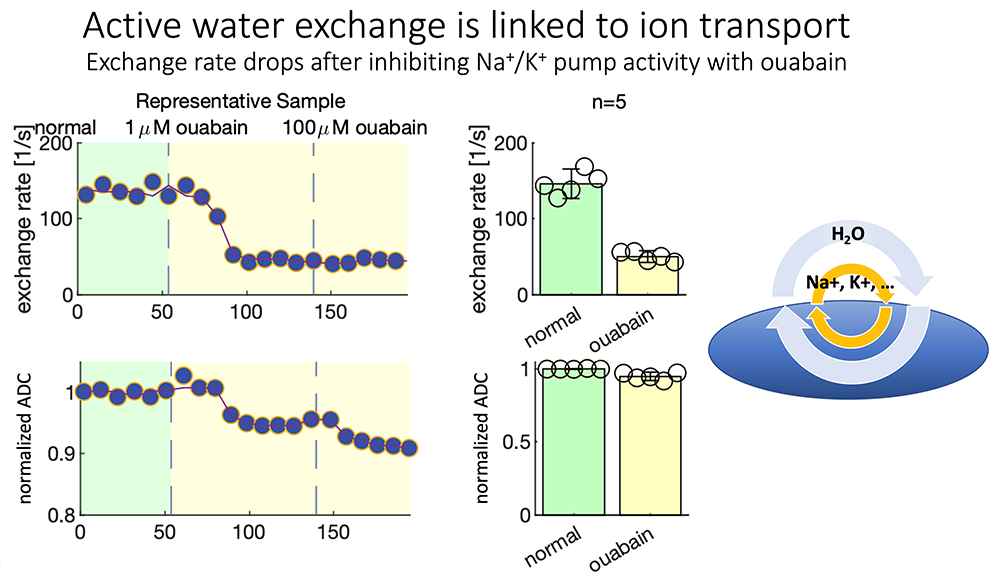
Figure 6. Steady-state water exchange provides a window on cell metabolism.
We are now able to measure steady-state water exchange without using exogenous dyes or contrast agents, but simply by watching water move among distinct compartments. The exchange rate, which characterizes the intensity of water exchange, can be modified by using a number of physical and chemical perturbations. Ouabain is a channel blocker that can down-regulate the operation of the Na-K ATPase membrane pump, and surprisingly, this is accompanied by a reduction in water exchange, adding support to the notion that water accompanies ions exchanged across cell membranes and could potentially detect the rate of ion exchange as a proxy for cell activity.
Patents
- Horkay F., Basser PJ. Composite gels and methods of use thereof, USPTO Patent number:20200254144, August 13, 2020
- Basser PJ. MRI Tractography Based Transit Time Determination for Nerve Fibers. USPTO Patent number: 20210270921. Publication date: September 2, 2021
Additional Funding
- “In vivo Brain Network Latency Mapping.” NIH BRAIN Initiative grant (no cost extension) 1-R24-MH-109068-01
- “Connectome 2.0: Developing the next generation human MRI scanner for bridging studies of the micro-, meso- and macro-connectome.” NIH BRAIN Initiative-funded 1U01EB026996-01
- “Neuroradiology/Neuropathology Correlation/Integration Core.” 309698-4.01-65310, (CNRM-89-9921)
Publications
- Saleem KS, Avram AV, Glen D, Yen CC, Ye FQ, Komlosh M, Basser PJ. High-resolution mapping and digital atlas of subcortical regions in the macaque monkey based on matched MAP-MRI and histology. Neuroimage 2021 245:118759.
- Avram AV, Saleem KS, Komlosh ME, Yen CC, Ye FQ, Basser PJ. High-resolution cortical MAP-MRI reveals areal borders and laminar substructures observed with histological staining. Neuroimage 2022 119653.
- Magdoom KN, Komlosh ME, Saleem K, Gasbarra D, Basser PJ. High resolution ex vivo diffusion tensor distribution MRI of neural tissue. Front Phys 2022 10:807000.
- Cai TX, Williamson NH, Ravin R, Basser PJ. Disentangling the effects of restriction and exchange with diffusion exchange spectroscopy. Front Phys 2022 23:805793.
- Benjamini D, Priemer DS, Perl DP, Brody DL, Basser PJ. Mapping astrogliosis in the individual human brain using multidimensional MRI. Brain 2022 awac298.
- Horkay F, Basser PJ. Hydrogel composite mimics biological tissues. Soft Matter 2022 18:4414–4426.
Collaborators
- Emilios Dimitriadis, PhD, Division of Bioengineering and Physical Science, NIBIB, Bethesda, MD
- Jack Douglas, PhD, NIST, Gaithersburg, MD
- R. Douglas Fields, PhD, Section on Nervous System Development and Plasticity, NICHD, Bethesda, MD
- Raisa Freidlin, PhD, Signal Processing and Instrumentation Section, CIT, NIH, Bethesda, MD
- Dario Gasbarra, PhD, University of Helsinki, Helsinki, Finland
- Mark R. Gilbert, MD, Neuro-Oncology Branch, Center for Cancer Research, NCI, Bethesda, MD
- Mark Haacke, PhD, Wayne State University School of Medicine, Detroit, MI
- Mark Hallett, MD, PhD, Human Motor Control Section, NINDS, Bethesda, MD
- Iren Horkayne-Szakaly, MD, Joint Pathology Center, Armed Forces Institute of Pathology, Washington, DC
- Beth Hutchinson, PhD, University of Arizona, Tucson, AZ
- Sara Inati, MD, Electroencephalography (EEG) Section, NINDS, Bethesda, MD
- Edward L. Mertz, PhD, Section on Physical Biochemistry, NICHD, Bethesda, MD
- Michael O'Donovan, MD, PhD, Developmental Neurobiology Section, NINDS, Bethesda, MD
- Evren Özarslan, PhD, Linköping University, Linköping, Sweden
- Sinisa Pajevic, PhD, Section on Critical Brain Dynamics, NIMH, Bethesda, MD
- Daniel Perl, MD, Uniformed Services University of the Health Sciences, Bethesda, MD
- Carlo Pierpaoli, MD, PhD, Section on Quantitative Medical Imaging, NIBIB, Bethesda, MD
- Dietmar Plenz, PhD, Section on Critical Brain Dynamics, NIMH, Bethesda, MD
- Tom Pohida, MS, Signal Processing and Instrumentation Section, CIT, NIH, Bethesda, MD
- Randall Pursley, MS, Signal Processing and Instrumentation Section, CIT, NIH, Bethesda, MD
- Roberto Romero, MD, D(Med)Sci, Perinatology Research Branch, NICHD, Detroit, MI
- Joelle Sarlls, PhD, In Vivo NMR Center, NINDS, Bethesda, MD
- Brain Development Cooperative Group, Various
Contact
For more information, email pjbasser@helix.nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/basser.