Exploring Genes and Signals Regulating Mammalian Hematopoiesis and New Approaches to Immunotherapy

- Paul E. Love, MD, PhD, Head, Section on Hematopoiesis and Lymphocyte Biology
- LiQi Li, MD, PhD, Staff Scientist
- Guillaume Gaud, PhD, Research Fellow
- Avik Dutta, PhD, Visiting Fellow
- Teri Hatzihristidis, PhD, Visiting Fellow
- Dalal El-Khoury, BS, Technician
- Jan Lee, BS, Technician
- Seeyoung Choi, PhD, Contract employee
- Layla Ahmadi, BS, Postbaccalaureate Fellow
Our research primarily focuses on the development of the mammalian hematopoietic system. A long-term area of interest for our laboratory is the study of signal-transduction molecules and pathways that regulate T lymphocyte maturation in the thymus. Currently, we are analyzing transgenic and conditional non-signaling mutants of T cell antigen receptor (TCR)–signaling subunits, made in our lab, to evaluate the importance of individual TCR–signaling chains and motifs at specific stages of T cell development and in mature T cells. In newer studies, we are investigating whether modification of TCR–signaling subunits can be used to enhance the tumoricidal activity of T cells for cancer treatment. Using gene profiling, we seek to identify proteins that are important for fine-tuning the T cell–signaling response in developing and mature T lymphocytes. In conjunction with checkpoint inhibitors for immunotherapy in humans, such molecules may also be targets to enhance anti-tumor activity. We also investigate the function of new T cell–specific proteins that we identified by subtraction-library screening. Our studies revealed a critical role for one of these proteins, called Themis, in T cell development by enhancing the TCR–signaling response in thymocytes. We found that another newly identified protein, Fbxl12, is important for regulating proliferation during early T cell development. Another area of investigation focuses on hematopoietic stem cells (HSCs) and early stages of T cell, B cell, and erythrocyte development. We initiated characterization of a protein (Ldb1) that is important for the generation and maintenance/self-renewal of HSCs, which revealed a critical function for Ldb1 as a key subunit of multimeric DNA–binding complexes in controlling the self-renewal/differentiation cell-fate decision in HSCs. Current work, which involves genome-wide screening by RNA-Seq and ChIP-Seq, seeks to extend our knowledge of the role of Ldb1 complexes in regulating gene transcription and to explore the function of such complexes in other hematopoietic lineages. For example, we have begun to investigate the importance of Ldb1 complexes for regulating self-renewal in immature thymocytes and in the induction of T cell acute lymphoblastic leukemia (T-ALL). Our results suggest that Ldb1 complexes represent potential therapeutic targets for the treatment of an aggressive form of childhood leukemia called early T progenitor T cell acute lymphoblastic leukemia (ETP-T-ALL).
T cell antigen receptor (TCR) signaling in thymocyte development
Much of our research in the past has focused on the role of TCR signal transduction in thymocyte development. Signal transduction sequences, termed immuno-receptor tyrosine-based activation motifs (ITAMs), are contained within four distinct subunits of the multimeric TCR complex (CD3-zeta, CD3-gamma, CD3-delta, and CD3-epsilon). Di-tyrosine residues within ITAMs are phosphorylated upon TCR engagement; their function is to recruit signaling molecules, such as protein tyrosine kinases, to the TCR complex, thereby initiating the T cell–activation cascade. Though conserved, ITAM sequences are non-identical, raising the possibility that the diverse developmental and functional responses controlled by the TCR may be partly regulated by distinct ITAMs through the recruitment of different effector molecules. We previously generated CD3-zeta–deficient and CD3-epsilon–deficient mice by gene targeting. We genetically reconstituted the mice with transgenes encoding wild-type or signaling-deficient (ITAM–mutant) forms of CD3-zeta and CD3-epsilon and characterized the developmental and functional consequences of the alterations for TCR signaling. We found that TCR–ITAMs are functionally equivalent but act in concert to amplify TCR signals and that TCR signal amplification is critical for thymocyte selection, the process by which potentially useful immature T cells are instructed to survive and differentiate further (positive selection) and by which potentially auto-reactive cells, which may cause autoimmune disease, are deleted in the thymus (negative selection).
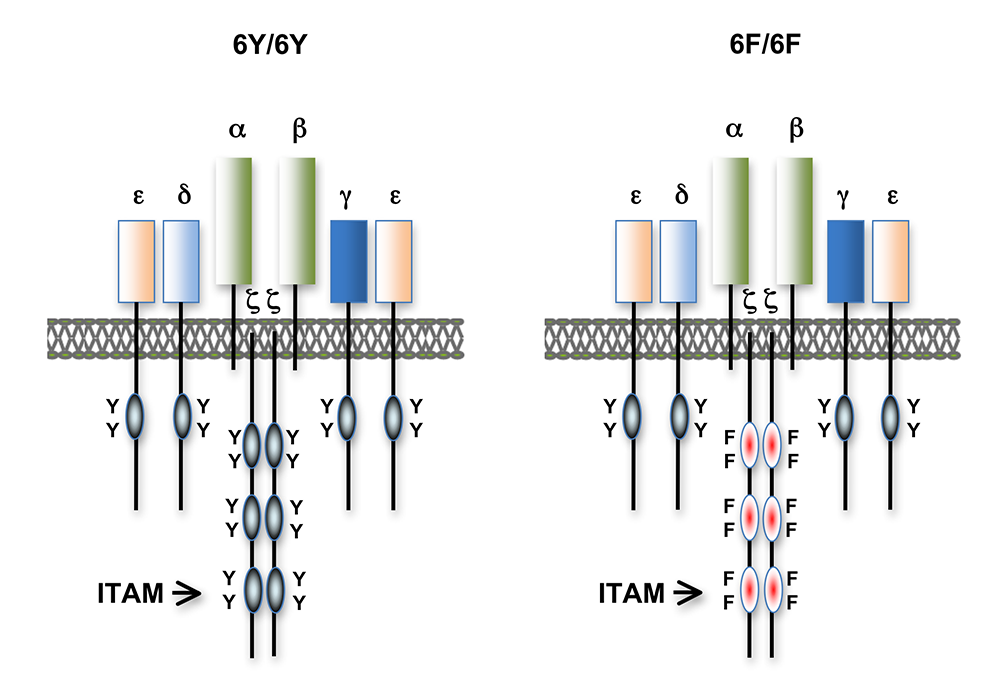
We also found that a complete complement of TCR–ITAMs is not required for most mature T cell effector functions. However, recent work demonstrated a requirement for ITAM multiplicity for the generation of T follicular helper cells, which are necessary for optimal B cell antibody responses. One possible explanation for the relatively mild phenotype observed in the TCR ITAM–reduced mice is that ITAM–mediated signal amplification is not required for most mature T cell activation responses; another is that some ITAMs perform more subtle, regulatory functions. To investigate this question further, we recently generated a TCR–zeta chain conditional knock-in mouse in which T cell development and selection can occur without attenuation of TCR signaling (i.e., in the presence of a wild-type 3-ITAM ‘6Y’ zeta chain), but in which mature, post-selection T cells may be induced to express TCRs containing signaling-defective (0-ITAM ‘6F’) zeta chains in lieu of wild-type zeta chains (Figure 1). Thus, mature T cell signaling should not be influenced by potential compensatory mechanisms that operate during T cell maturation, and T cells in such mice should be faithful indicators of the role of multiple TCR ITAMs in mediating specific, mature T cell responses. We confirmed that the knock-in zeta locus functions as predicted. We next evaluated the effect of late ‘switching’ from 6Y zeta to 6F zeta in mature T cells generated with wild-type 6Y zeta–containing TCRs and found that the phenotype was identical to germline inactivation of zeta ITAMs, demonstrating that compensation does not explain the mild phenotype of zeta 6F mice. Unexpectedly, we also discovered an inhibitory role for zeta ITAM signaling in response to weak (low-affinity) antigens. Strikingly, inactivation of the zeta ITAMs resulted in enhanced TCR signaling and enhanced T cell effector functions when the TCR is engaged by low-affinity ligands, but zeta ITAMs contributed positively to signaling by high-affinity ligands. This revealed a dual (activating and inhibitory) function for zeta ITAMs in TCR signaling depending on the affinity of the TCR–ligand interaction. Given that most tumor-specific antigens are low affinity and that this property limits current TCR–based approaches to tumor immunotherapy, we explored the effect of zeta ITAM inactivation on T cell–tumoricidal activity. Notably, we found that inactivation of zeta ITAMs markedly enhanced T cell–tumoricidal activity against low-affinity tumor antigens. Such experiments should provide information relevant to the design of engineered tumor antigen–specific TCRs and possibly chimeric antigen receptor T cells (CAR T cells), which are currently configured to express zeta ITAM–signaling module(s).
Figure 1. T cell antigen receptors expressed in 6Y/6Y and 6F/6F knock-in mice
Subunit composition of the T cell antigen receptors in 6Y/6Y and 6F/6F mice. 6Y/6Y mice express wild-type zeta chain dimers with functional ITAM signaling motifs that contain two tyrosine (Y) residues. 6F/6F mice express mutant zeta chain dimers in which the ITAM tyrosines have been changed to phenylalanine (F).
Identification and characterization of TCR–tuning proteins that may serve as targets for immunotherapy
We extended our analysis of TCR–signaling subunits to other molecules that participate in or influence the TCR–signaling response. The cell-surface protein CD5 negatively regulates TCR signaling and functions in thymocyte selection. Examination of CD5 expression during T cell development revealed that surface levels of CD5 are regulated by TCR–signal intensity and by the affinity of the TCR for self-peptide ligands in the thymus that mediate selection. To determine whether the ability to regulate CD5 expression is important for thymocyte selection, we generated transgenic mice that constitutively express high levels of CD5 throughout development. Over-expression of CD5 significantly impaired positive selection of some thymocytes (those that would normally express low levels of CD5) but not of others (those that would normally express high levels of CD5). The findings support a role for CD5 in modulating TCR signal transduction, thereby influencing the outcome of thymocyte selection. Current studies center on identifying the mechanism by which CD5 inhibits TCR signaling and on determining whether the protein's regulated expression during development is important for preventing autoimmunity. For that purpose, we generated a conditional CD5–deletion mouse in which CD5 expression can be removed before, during, or after T cell development. The ability of individual thymocytes to regulate CD5 expression represents a mechanism for ‘fine tuning’ the TCR–signaling response during development so that the integrated signaling response can be adjusted to permit T cell functional competency without causing autoimmunity. Reasoning that, in addition to CD5, other molecules participate in TCR tuning, we initiated microarray-based screening for genes differentially expressed in developing T cells under conditions of high- or low-affinity TCR interactions. We identified several genes from this screen for further study, including CD6, a surface receptor that is structurally similar to CD5, and we are validating their function as tuning molecules. Given that the molecules regulate TCR signaling, they represent potential autoimmune-disease susceptibility markers and potential targets for treatment of patients with cancer or autoimmune disease, similar to current ‘checkpoint inhibitor’ therapies that are based on blocking the function of the induced inhibitory molecules PD-1 and CTLA-4. Experiments are under way to investigate this translational potential.
Identification and characterization of Themis, a novel protein required for T cell development
Using a subtractive cDNA library–screening approach, we identified Themis, now known as Themis1, a novel T cell–specific adapter protein (Figure 2). To investigate the function of Themis1 in T cell signaling and development, we generated Themis1 knock-down cell lines, Themis1 knock-out mice (conventional and conditional), and Themis1 transgenic mice. Analysis of the effects of modulating Themis1 expression revealed a critical role for the protein in late T cell development. In a collaboration with Richard Cornall, we also investigated the phenotype of Themis2 knockout mice generated in our lab. Our results identified an important role for Themis2 in facilitating B cell activation by low-avidity, but not high-avidity, B cell receptor (BCR)–antigen interactions.
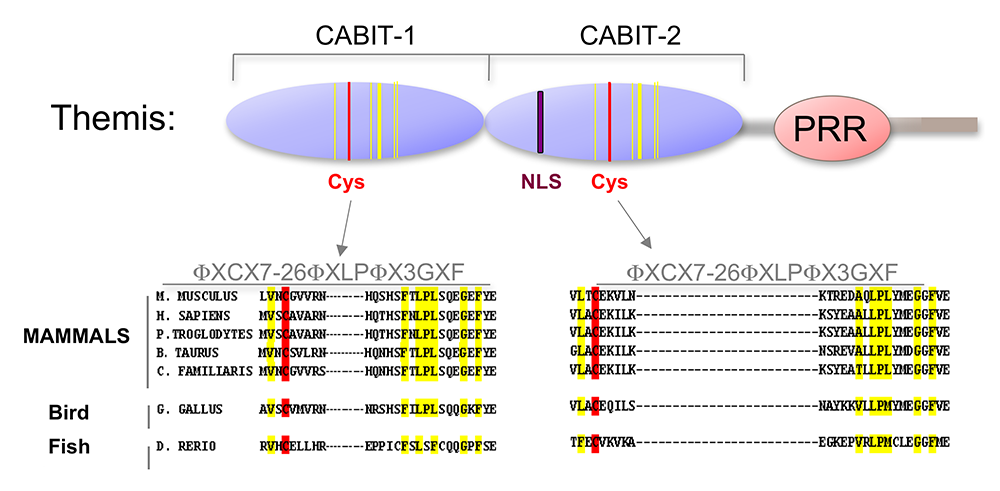
In the past few years, we focused on determining the molecular function of Themis1. Themis1, Themis2, and a large family of related metazoan proteins contain a novel globular domain of unknown function called the CABIT (cysteine-containing, all beta in Themis) module (Figure 2). Using cell transfection and biochemical and protein-assay techniques, we determined that CABIT modules bind to the catalytic domain of SHP-1, a key hematopoietic protein tyrosine phosphatase. In the presence of reactive oxygen species (ROS), which are generated in activated T cells, Themis1, via its CABIT module, promoted oxidation of the SHP-1 catalytic cysteine and therefore inactivated SHP-1. The CABIT modules from all five mammalian Themis-family proteins also inhibited SHP-1, indicating that this activity was common to the CABIT module. Given that SHP-1 is an inhibitory phosphatase that functions to dampen TCR signaling by de-phosphorylating several targets, including protein tyrosine kinases, the finding established an activating function for Themis1 in cell signaling through its ability to bind to and inhibit SHP-1. Interestingly, Themis1 is highly expressed in developing thymocytes at the stage at which they undergo positive selection. It had been known for years that thymocytes are more sensitive to TCR stimulation than are mature T cells, but the reason for this sensitivity was unknown. The function of Themis1, together with its high expression in thymocytes, provides an explanation for the sensitivity of thymocytes to TCR signaling. By showing that deletion of the gene encoding SHP-1 rescues T cell development in Themis1–/– mice, we confirmed that the primary role of Themis1 is to inhibit SHP-1. In addition to identifying the function of CABIT modules, our results provide insight into the role of other metazoan CABIT–containing proteins (which number in the hundreds). Our ongoing studies are focusing on further characterization of CABIT proteins, including the intestine-specific mouse protein Themis3, and determining their role in development and their possible involvement in human disease.
Figure 2. Themis is highly conserved in vertebrates.
Themis contains two novel CABIT domains, each with a conserved cysteine (red) and conserved flanking residues (yellow), a nuclear localization signal (NLS), and a proline-rich region (PRR).
Role of the F-box protein Fbxl12 in thymocyte development
A major aspect of the T cell maturation process is the precise regulation of cell proliferation. Rather than being a shared property among all or most developing thymocytes, proliferation is strictly limited to two stages during early development. The initial proliferative phase is driven by thymus-expressed cytokines, the second coincides with ‘beta selection’ (i.e., is initiated in cells that have productively rearranged the TCRbeta chain and express a signaling complex called the pre–TCR). The proliferative burst that accompanies beta selection is estimated to result in a 100–200 fold expansion and is essential for further differentiation and for maximizing TCR diversity. Previous work showed that beta selection–associated proliferation requires concurrent signals by the pre–TCR and Notch receptors, but how these signals induce cell-cycle progression and why they need to be coordinated has remained unclear. Initiation of proliferation in beta-selected thymocytes requires the ubiquitin-mediated degradation of the cyclin-dependent kinase inhibitor Cdkn1b, which acts to prevent cell-cycle progression. In a recent study, we examined the molecular control of beta selection–associated proliferation. We confirmed prior findings that Cdkn1b degradation is induced by an SCF E3 ubiquitin ligase that contains the ligand-recognition subunit Fbxl1. Deletion of Fbxl1 partially blocked beta selection–associated proliferation, a defect that was rescued by co-deletion of Cdkn1b. We identified a new F-box protein, Fbxl12, that is highly expressed in thymocytes. We found that Fbxl12 also functions as an SCF E3 ligase subunit, which, like Fbxl1, directs Cdkn1b degradation. The phenotype of Fbxl12–deficient mice generated in our lab was strikingly similar to that of Fbxl1–deficient mice, and deletion of both Fbxl1 and Fbxl12 resulted in a severe block in beta selection–associated proliferation, indicating that Fbxl1 and Fbxl12 act in concert to regulate thymocyte proliferation. Interestingly, we found that Fbxl1 expression is induced by Notch signaling, whereas Fbxl12 expression is induced by pre–TCR signaling. Both Fbxl1 and Fbxl12 are required for thymocyte proliferation; thus, their selective regulation by Notch and the pre–TCR, respectively, provides an explanation for why concurrent Notch and pre–TCR signaling are necessary for cell-cycle progression and proliferation at the beta-selection checkpoint.
Role of Ldb1 transcription complexes in hematopoiesis and in T cell acute lymphoblastic leukemia (T-ALL)
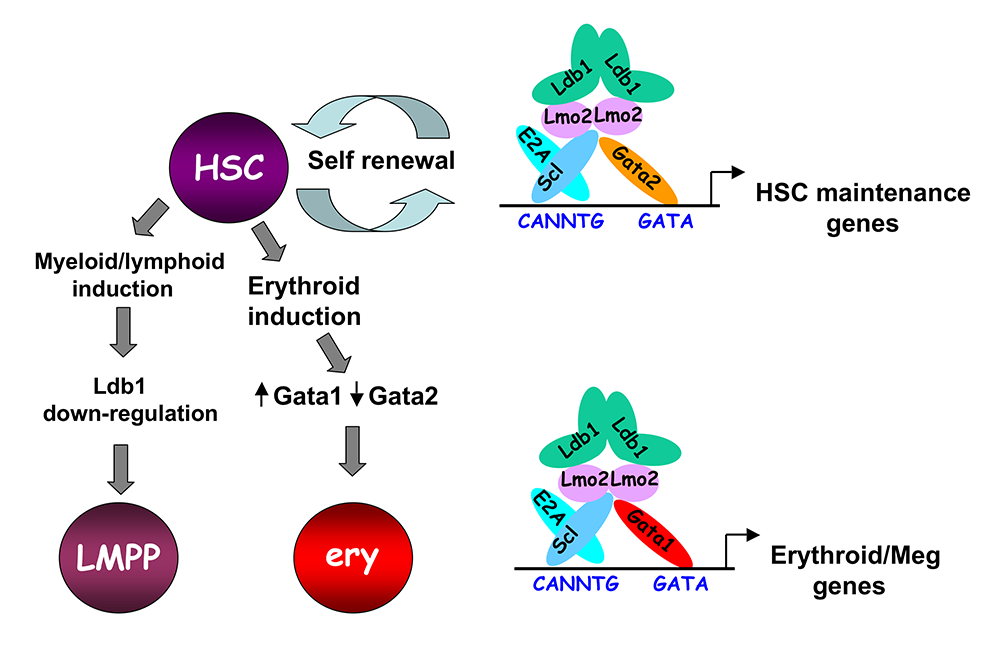
Lim domain binding protein-1 (Ldb1) is a ubiquitously expressed nuclear protein that contains a LIM–zinc finger protein–interaction motif and a dimerization domain. In hematopoietic cells, Ldb1 functions by interacting with and/or recruiting specific partners (including the LIM–only protein Lmo2 and the transcription factors Lyl1 or Tal1 and Gata1 or Gata2) to form multi-molecular transcription complexes (Figure 3). Within the hematopoietic lineage, expression of Ldb1 is highest in progenitor cells, which include hematopoietic stem cells (HSCs). We initially investigated the role of Ldb1 in hematopoiesis by following the fate of Ldb1–/– embryonic stem cells (ESCs) in mouse blastocyst chimeras and by conditional, stage-specific deletion of Ldb1 in HSCs. We found that Ldb1 is not required for ESC maintenance but is required for HSC maintenance. More recent data indicate that the loss of Ldb1–/– HSCs results from differentiation rather than cell death. We performed a genome-wide screen for Ldb1–binding sites using ChIP-Seq. Analysis of the ChIP-Seq data revealed that Ldb1 complexes bind at the promoter or at regulatory sequences near a large number of genes known to be required for HSC maintenance. Examination of the function of Ldb1 in cell lineages downstream of HSCs identified an essential function in the erythroid lineage but not in myeloid cells or lymphoid cells. Interestingly, ChIP-Seq analysis of Ldb1 DNA–binding complexes demonstrated that Ldb1 complexes in HSCs contain the transcription factor Gata2, whereas Ldb1 complexes in erythroid progenitors contain Gata1 (which is highly expressed in the erythroid lineage). The results indicate that multimeric Ldb1 transcription complexes have distinct functions in the hematopoietic system depending on their subunit composition, with Gata2–containing complexes regulating expression of HSC–maintenance genes and Gata1 complexes regulating expression of erythroid-specific genes (Figure 3). Current studies aim to determine how Ldb1 complexes regulate gene expression and the role of Ldb1 dimerization in mediating long-range promoter-enhancer interactions in hematopoietic cells. In addition, we are investigating a potential role for Ldb1 in regulating self-renewal of T cell progenitors in the thymus.
Figure 3. Model of Ldb1 function in the hematopoietic lineage
Ldb1 forms a multimeric DNA–binding complex in hematopoietic cells with the adapter Lmo2 and the transcription factors Scl and Gata1 or Gata2. In hematopoietic stem cells (HSCs), in which Gata2 is highly expressed, Ldb1-Lmo2-Scl-Gata2 complexes positively regulate expression of HSC maintenance genes. Differentiation of HSCs to the myeloid or lymphoid lineage (LMPP) is triggered by downregulation of Ldb1 complexes, whereas commitment to the erythroid lineage (ery) is triggered by induction of Gata1 and downregulation of Gata2, resulting in the formation of an Ldb1-Lmo2-Scl-Gata1 complex, which positively regulates expression of erythroid-specific genes.
Acute lymphoblastic leukemias are the most common type of cancer in children. T cell acute lymphoblastic leukemia (T-ALL) results from oncogenic transformation of immature T cell progenitors (thymocytes). Mouse models of T-ALL have been generated, and one of the most informative is the Lmo2–transgenic (Lmo2-tg) mouse, which expresses high levels of the nuclear adapter Lmo2 in thymocytes. The model closely mimics a prevalent type of human T-ALL, which is associated with chromosomal mutations that result in increased expression of LMO2. We recently reported that overexpression of Lmo2 in mouse thymocytes induces T-ALL at two distinct stages of development (an early ‘ETP’ stage and a later ‘DN3’ stage). Notably, human T-ALLs can also occur at two similar stages of thymocyte maturation. The most immature forms of T-ALL in Lmo2-tg mice and in humans express high levels of the transcription factor Hhex and are designated early T progenitor (ETP) T-ALL, whereas later-stage tumors are low in Hhex but express high levels of more mature markers of T cell development, including Notch1, Dtx1, Ptcra, and Hes1. Lmo2 functions as a subunit of the multimeric Ldb1–nucleated DNA–binding complexes described above. We found that normal ETP thymocyte progenitor cells express the same Ldb1 complex subunits as are present in HSCs and that ETPs exhibit HSC characteristics, including the self-renewal potential. ETPs in Lmo2-tg mice appear to be ‘locked’ into a pattern of perpetual self-renewal and are refractory to normal inductive signals that promote further differentiation. Hhex is a target of Ldb1 complexes in HSCs and ETPs, a result that strongly suggests that Ldb1 complexes are responsible for the aberrant self-renewal in Lmo2-tg mice that predisposes to oncogenesis. We hypothesized that Ldb1 complexes regulate self-renewal in ETPs as well as in HSCs. Lmo2 is normally down-regulated when thymocytes undergo T lineage commitment, suggesting that extinguishing expression of Lmo2 (and by extension, Ldb1 complexes) is important for T cell differentiation and that failure to do so predisposes to oncogenesis via ‘second-hit’ transforming events.
In RNA-Seq gene expression experiments, we found that the RNA–expression signatures of Lmo2-tg immature thymocytes and HSCs are very similar, consistent with the notion that Lmo2 overexpression ‘freezes’ cells in a stem-cell self-renewal state. To determine whether Ldb1 complexes are in fact required for ETP self-renewal and to explore the genes regulated by these complexes, we conditionally deleted Ldb1 in Lmo2-tg mice. We found that Ldb1 is required for Lmo2-tg–induced thymocyte self-renewal and T-ALL induction, indicating that Lmo2 overexpression promotes T-ALL by functioning as a subunit of Ldb1 complexes. Currently, we are addressing several key questions, including whether, as predicted, Ldb1, and by extension Ldb1 complexes, regulate expression of genes that control a self-renewal genetic program in ETPs and whether Ldb1 complexes are necessary for the transcriptional/developmental effects of Lmo2. We are also determining the subunit structure and binding sites of Ldb1 complexes expressed in Lmo2-tg thymocytes. We anticipate that our results will provide insights into the mechanisms controlling T-ALL oncogenesis in humans and may thus provide new therapeutic avenues for the treatment of this devastating pediatric disease.
Additional Funding
- NICHD Director's Award
- NIH IRF Award to Avik Dutta
Publications
- Dutta A, Zhao B, Love PE. New insights into beta-selection. Trends Immunol 2021 42:735–750.
- Stamos DB, Clubb LM, Mitra A, Chopp L, Nie J, Ding Y, Das A, Venkataganesh H, Lee J, El-Khoury D, Li L, Bhandoola A, Bosselut R, Love PE. The histone demethylase Lsd1 regulates multiple repressive gene programs during T cell development. J Exp Med 2021 218(12):e20202012.
- Yang C, Blaize G, Marrocco R, Rouquié N, Bories C, Gador M, Mélique S, Joulia E, Benamar M, Dejean AS, Daniels-Treffandier H, Love PE, Fazilleau N, Saoudi A, Lesourne R. THEMIS enhances the magnitude of normal and neuroinflammatory type 1 immune responses by promoting TCR-independent signals. Sci Signal 2022 15(742):eabl5343.
- Schuster C, Kiaf B, Hatzihristidis T, Ruckdeschel A, Nieves-Bonilla J, Ishikawa Y, Zhao B, Zheng P, Love PE, Kissler S. CD5 controls gut immunity by shaping the cytokine profile of intestinal T cells. Front Immunol 2022 13:906499.
- Gao Y, Zamisch M, Vacchio M, Chopp L, Ciucci T, Paine EL, Lyons GC, Nie J, Xiao Q, Zvezdova E, Love PE, Vinson CR, Jenkins LM, Bosselut R. NuRD complex recruitment to Thpok mediates CD4+ T cell lineage differentiation. Sci Immunol 2022 7(72):eabn5917.
- Zhang M, Lin X, Yang Z, Li X, Zhou Z, Love PE, Huang J, Zhao B. Metabolic regulation of T cell development. Front Immunol 2022 13:946119.
Collaborators
- Gregoire Altan-Bonnet, PhD, Center for Cancer Research, NCI, Bethesda, MD
- Cheryl Arrowsmith, PhD, University of Toronto, Toronto, Canada
- Avinash Bhandoola, MB, PhD, Laboratory of Genome Integrity, Center for Cancer Research, NCI, Bethesda, MD
- Remy Bosselut, MD, PhD, Laboratory of Immune Cell Biology, NCI, Bethesda, MD
- Utpal P. Davé, MD, Vanderbilt-Ingram Cancer Center, Vanderbilt University, Nashville, TN
- Richard J. Cornall, FRCP, FMedSci, Nuffield Department of Medicine, University of Oxford, Oxford, United Kingdom
- Lauren Ehrlich, PhD, The University of Texas at Austin, Austin, TX
- James Gulley, MD, PhD, Center for Cancer Research, NCI, Bethesda, MD
- Christian Hinrichs, MD, Rutgers Cancer Institute, New Brunswick, NJ
- Aravind Iyer, PhD, Protein and Genome Evolution Research Group, NLM/NCBI, Bethesda, MD
- Park Jung-Hyun, PhD, Experimental Immunology Branch, Center for Cancer Research, NCI, Bethesda, MD
- Stephan Kissler, PhD, Joslin Diabetes Center, Harvard Medical School, Boston, MA
- Renaud Lesourne, PhD, INSERM, Toulouse, France
- Dorian McGavern, PhD, Viral Immunology and Intravital Imaging Section, NINDS, Bethesda, MD
- Keiko Ozato, PhD, Section on Molecular Genetics of Immunity, NICHD, Bethesda, MD
- Rita Perlingeiro, PhD, University of Minnesota Medical School, Minneapolis, MN
- Karl Pfeifer, PhD, Section on Epigenetics, NICHD, Bethesda, MD
- Alfred Singer, MD, Experimental Immunology Branch, Center for Cancer Research, NCI, Bethesda, MD
- Nevil Singh, PhD, University of Maryland School of Medicine, Baltimore, MD
- Naomi Taylor, MD, PhD, Pediatric Oncology Branch, Center for Cancer Research, NCI, Bethesda, MD
- Juan Carlos Zúñiga-Pflücker, PhD, University of Toronto, Toronto, Canada
Contact
For more information, email lovep@mail.nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/love.