Neuronal Circuits Controlling Behavior: Genetic Analysis in Zebrafish

- Harold Burgess, PhD, Head, Section on Behavioral Neurogenetics
- Tripti Gupta, PhD, Staff Scientist
- Ashwin A. Bhandiwad, PhD, Postdoctoral Fellow
- Jennifer Panlilio, PhD, Postdoctoral Fellow
- Svetlana Semenova, PhD, Visiting Fellow
- Reid Doctor, BSc, Graduate Student
- Daniel Bazan, BSc, Postbaccalaureate Fellow
- Grace Biddle, BSc, Postbaccalaureate Fellow
- George Holmes, BSc, Postbaccalaureate Fellow
- Katelyn Robertson, Special Volunteer
- Jennifer L. Sinclair, MSc, Zebrafish Technician
The Section on Behavioral Neurogenetics studies how, under diverse environmental contexts, the nervous system selects appropriate behavioral responses to sensory information in a way that best satisfies internal motivational objectives. We use the larval zebrafish as a model because its brain exhibits the basic architecture of the vertebrate brain but is much less complex than the mammalian brain. Despite the relative simplicity of their nervous system, zebrafish have a sophisticated repertoire of sensory-guided and internally driven behaviors. Furthermore, the optical clarity of the embryo facilitates visualization of individual neurons and their manipulation with genetic techniques. Behavior in larvae is innate and thus exhibits minimal variability between fish. Subtle alterations in behavior can therefore be robustly measured, making it possible to quickly assess the contribution of identified neurons to a variety of motor behaviors.
We focus on two aspects of behavioral regulation: the neuronal mechanisms by which sensory context regulates behavioral decisions and the pathways that sustain changes in behavioral state. Neuronal connections that allow the brain to integrate sensory and internal-state information are established through genetic interactions during development, and are frequently disrupted by gene mutations associated with neuro-developmental disorders. We can therefore use discoveries about sensorimotor integration pathways to understand how human disease genes disrupt brain development. To support these objectives, we develop new genetic tools and behavioral assays to probe the nexus between neuronal function and behavior at single-cell resolution.
Neuronal pathways for auditory sensory processing
Startle responses are rapid reflexes that are triggered by sudden sensory stimuli, and which help animals defend against, or escape from, potentially threatening stimuli. In both fish and mammals, startle responses are initiated by giant reticulospinal neurons in the medulla, which receive short-latency sensory input from diverse sensory modalities. Although highly stereotyped, startle responses are nevertheless modulated by sensory context and behavioral state and are therefore an excellent system in which to understand how such information is integrated for behavioral choice. In mammals, including humans, the startle response to a strong auditory stimulus can be inhibited by pre-exposure to a weak acoustic ‘prepulse,’ a form of startle modulation, termed prepulse inhibition, that is diminished in several neurological conditions. Previously, we showed that in zebrafish, as in mammals, several distinct cellular mechanisms mediate prepulse inhibition, depending on the time interval between the prepulse and the startle stimulus, with NMDA–receptor signaling playing a key role for intervals greater than 100 ms. Our work on resolving the core neuronal pathway that mediates prepulse inhibition provides a basis for probing how gene mutations linked to neuro-developmental disorders disrupt sensory processing. NMDA–receptor mutations have been linked to both autism-spectrum disorders and schizophrenia. Working collaboratively with the Sirotkin laboratory, we demonstrated that mutations in the NMDA–receptor subunit grin2B disrupt neural development [Reference 1].
Neural mechanisms for behavioral-state control
Over the course of the day, motivational goals change in response to both internal and external cues. At any given moment, an individual’s behavioral state strongly influences decisions on how to interact with the environment. A major goal in neuroscience is to identify the neural systems that maintain short-term behavioral states and to determine how they interact with central mechanisms for behavioral choice.
While studying defensive responses to auditory stimuli, we noted that cues of about an order of magnitude larger than those needed to provoke startle responses drove an unexpected freezing behavior, especially when repeatedly presented over a short period of time. In many species, overwhelming stimuli elicit a behavior known as tonic immobility, an ultimate response to inescapable threat. In humans, the experience of tonic immobility correlates with development of the post-traumatic stress disorder, yet very little is known about its underlying neural basis. Leveraging our library of Gal4 (a tool to modulate gene activity)–transgenic lines, we performed a screen to discover neurons that are required for such behavior in zebrafish. We isolated a cluster of neurons in the prepontine tegmentum that are necessary for sustained immobility after an intense auditory stimulus (Figure 1). By manually isolating and performing RNA-Seq on these neurons, we found that they express several stress-associated neuropeptides, including markers that make them likely homologs of part of the mammalian parabrachial complex, an area recently implicated in responses to noxious stimuli. Our screen also demonstrated a central role for cerebellar signaling in tonic immobility, and we found a direct projection from Purkinje neurons to the prepontine neurons, similar to recent work showing that a subset of Purkinje neurons in mammals also directly project to the parabrachial nucleus. The study identified, for the first time, a cellular pathway that mediates tonic immobility and suggests that the parabrachial has a deep evolutionary history in mediating defensive behavior [Reference 2].
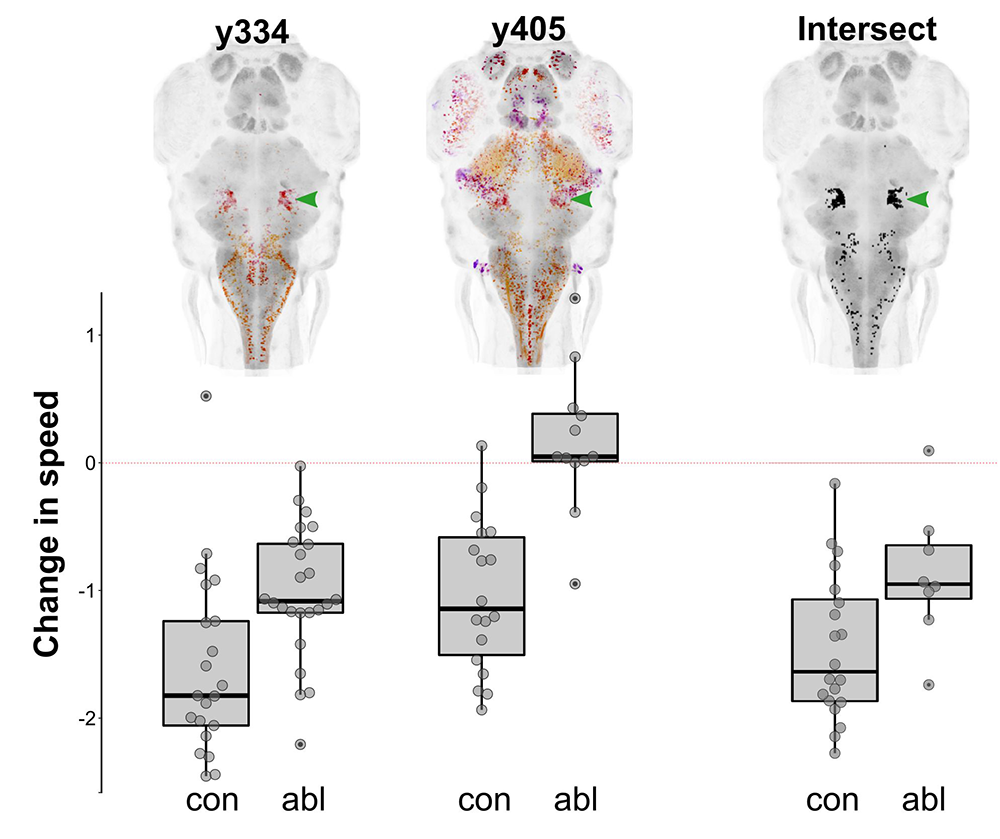
Figure 1. Prepontine tegmental neurons mediate tonic immobility in zebrafish.
Depth-coded projections of Gal4 expression in two lines (y334 and y405) that label neurons whose ablation interferes with the reduction in spontaneous swimming in response to an intense auditory stimulus (bar graphs). The computational intersectional expression of y334 and y405 is shown, revealing a common cluster of neurons in the prepontine tegmentum (green arrowhead). Laser ablation of the prepontine cluster diminishes immobility in response to an intensity stimulus.
Zebrafish models of neuro-developmental disorders
We collaborate widely with clinicians to generate and characterize zebrafish models for mutations discovered in humans (often through exome-sequencing) that are likely to have a neuro-developmental origin. We use the CRISPR/Cas9 system to generate lesions in zebrafish genes that are homologous to those disrupted in disorders. We then apply behavioral analysis, transcriptomics, and voxel-based morphometry as part of a broad phenotyping strategy. To promote rigorous use of zebrafish neurological disease models, we wrote a critical review outlining advantages and limitations of the zebrafish system [Reference 3 & 4].
Our work with brain morphometry arose from earlier studies in which we generated several hundred new Gal4 and Cre lines in order to provide genetic accessibility to neurons of interest. A unique feature of brain imaging in zebrafish is the ability to visualize the total architecture of the brain while simultaneously recording the position and morphology of every constituent labeled neuron. To make these transgenic lines accessible to the broader research community, we performed whole-brain imaging for each line, then registered the image of each line to the same reference brain (Figure 2). In collaboration with Nicholas Polys, we then developed an online brain atlas that enables researchers to quickly visualize the larval brain and locate transgenic lines to aid experiments. Such powerful visualization tools facilitate integrated analysis of reconstructed neuronal morphology in the context of the three-dimensional anatomy of the brain. Then, in order to build a brain atlas, we optimized a protocol that permits highly precise brain registration [Reference 5].
Figure 2. Whole brain registration in zebrafish
A. Brain registration corrects for both technical variability during imaging and biological variability between samples. Three larval zebrafish brains, pseudocolored red, green and blue, were imaged. When superimposed, it is apparent that they do not precisely align. Following registration, the same three brains are closely coordinated, even to the extent that the Mauthner cells, an identifiable bilateral cell in the hindbrain of each brain, become closely matched.
B. Brain registration allows transgene expression patterns from different individuals to be mapped onto the same reference brain coordinate system, so that they can be compared in three dimensions. In this example, four transgenic lines (pseudo-colored) were separately imaged then, following registration, simultaneously visualized.
We showed that such high precision of alignment permits statistically robust whole-brain analysis of neuronal composition and morphology in zebrafish mutant models, pinpointing brain regions with changes that are difficult to detect visually (Figure 3). The technique can be applied to almost any zebrafish neuro-developmental model, thereby enabling robust and quantitative detection of subtle changes in brain structure or composition. We used this method to test brain structure and composition in zebrafish that carry mutations in genes that are homologous to human genes known to be disrupted in a variety of neuro-developmental disorders, including autism and intellectual disability. Through this work, we aim to provide insight into the fundamental molecular and cellular processes associated with each disorder.
Figure 3. Pipeline for brain morphometry in zebrafish
A. Control and mutant brains are scanned and co-registered.
B. We calculate p-value maps of intensity and size differences at each voxel.
C. Clusters of significant voxels reveal local changes independent of anatomical boundaries.
D–E. Comparing mean values in neuroanatomically defined regions provides strong statistical power to detect subtle changes in brain composition or structure in models of neuro-developmental disorders.
Publications
- Zoodsma J, Keegan E, Bhandiwad A, Napoli AJ, Burgess HA, Sirotkin H, Wollmuth L. Disruption of grin2B, an ASD-associated gene, produces social deficits in zebrafish. Mol Autism 2022 13:38.
- Bhandiwad AA, Chu N, Semenova SA, Burgess HA. A cerebellar-prepontine circuit for tonic immobility triggered by inescapable threat. Sci Adv 2022 8:eabo0549.
- Burgess HA, Burton EA. A critical review of zebrafish neurological disease models – 1. The premise: neuroanatomical, cellular, and genetic homology, and experimental tractability. Oxford Open Neuroscience 2023 kvac018.
- Burton EA and Burgess HA. A critical review of zebrafish neurological disease models – 2. Application: functional and neuroanatomical phenotyping strategies and chemical screens. Oxford Open Neuroscience 2022 kvac019.
- Bhandiwad AA, Gupta T, Subedi A, Heigh V, Holmes GA, Burgess HA. Brain imaging and registration in larval zebrafish. Zenodo 2022 10.5281/zenodo.7312001.
Collaborators
- Edward A. Burton, MD, DPhil, FRCP, Pittsburgh Institute for Neurodegenerative Diseases, University of Pittsburgh, Pittsburgh, PA
- Andy Golden, PhD, Laboratory of Biochemistry and Genetics, NIDDK, Bethesda, MD
- Todd S. Macfarlan, PhD, Section on Mammalian Epigenome Reprogramming, NICHD, Bethesda, MD
- Reza Maroofian, PhD, Institute of Neurology, University College, London, UK
- Anne O'Donnell Luria, MD, PhD, Mendelian Genomics Research Center, Broad Institute of MIT and Harvard, Cambridge, MA
- Nicholas F. Polys, PhD, Department of Computer Science, Virginia Tech, Blacksburg, VA
- Howard Sirotkin, PhD, Stony Brook University, Stony Brook, NY
- Lonnie P. Wollmuth, PhD, Stony Brook University, Stony Brook, NY
Contact
For more information, email haroldburgess@mail.nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/burgess.