Signaling and Secretion in Neuroendocrine Cells

- Stanko S. Stojilkovic, PhD, Head, Section on Cellular Signaling
- Stephanie Constantin, PhD, Staff Scientist
- Rafael M. Previde, PhD, Visiting Fellow
- Kosara Smiljanic, PhD, Visiting Fellow
- Srdjan J. Sokanovic, PhD, Visiting Fellow
The main goal of the research in our Section is to examine cell-signaling cascades, gene expression, and hormone secretion in neuroendocrine cells from the hypothalamus and pituitary gland during development. We place special emphasis on the characterization of individual cells, using fluorescence imaging, patch-clamp recordings, simultaneous membrane potential/calcium and current/calcium recordings, electrophysiological and imaging recordings of single-cell exocytic events, single-cell RNA sequencing (scRNA-Seq), and single-cell quantitative reverse transcription polymerase chain reaction (RT-PCR). Our recent and ongoing work has focused on the following: signaling, transcription, and secretion in the pituitary gland specific for age, sex, and tissue structure; pituitary cell heterogeneity, reflecting their postnatal gene expression; the role of 1-phosphatidylinositol 4-kinases and protein receptor tyrosine phosphatase N2 in postnatal proliferation and maintenance of pituitary lineages; and cell-specific electrical activity and exocytic pathways. Current and proposed studies depend in part on the use of equipment in NICHD's Microscopy and Imaging Core Facility and the Molecular Genomics Core Facility.
Role of PI4-kinase alpha in calcium signaling and prolactin secretion
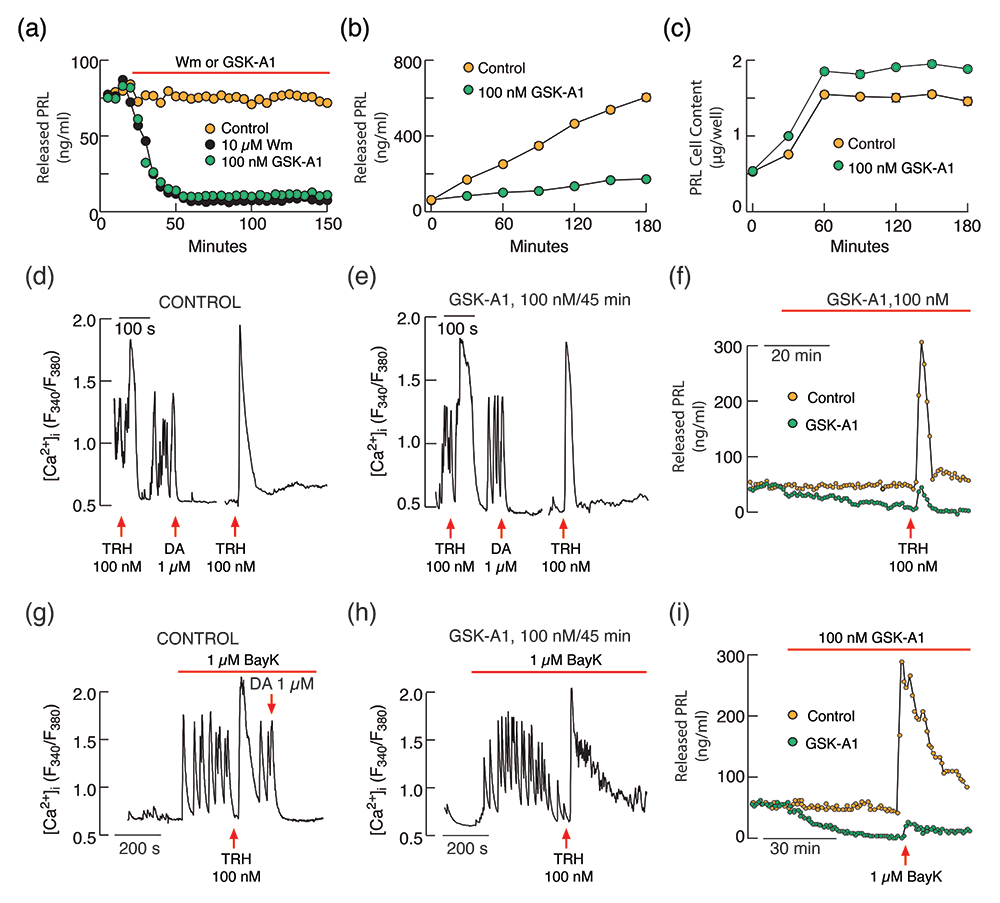
We continue investigations on genes expressed in pituitary cells and their roles in signaling and hormone secretion, focusing on the transcriptome profiles of secretory and non-secretory cell types, using scRNA-Seq of freshly dispersed pituitary cells from adult female rats. Among others, the studies revealed that all hormone-producing cells express the phosphatidylinositol (PI) kinase genes Pi4ka, Pi4kb, Pi4k2a, Pi4k2b, Pip5k1a, Pip5k1c, and Pik3ca, as well as Pikfyve and Pip4k2c. In a recent study, we analyzed the contribution of phosphatidylinositol kinases to calcium-driven prolactin (PRL) release in pituitary lactotrophs: PI4Ks, which control PI4P production; PIP5Ks, which synthesize PI(4,5)P2 by phosphorylating the D-5 position of the inositol ring of PI4P; and PI3KCs, which phosphorylate PI(4, 5)P2 to generate PI(3,4,5)P3. We used common and PIK–specific inhibitors to evaluate the strength of calcium-secretion coupling in rat lactotrophs. Wortmannin, a PI3K and PI4K inhibitor, but not LY294002, a PI3K inhibitor, blocked spontaneous action potential–driven PRL release with a half-time of about 20 minutes when applied in 10 μM concentration, leading to accumulation of intracellular PRL content. Wortmannin also inhibited the increase in PRL release by high potassium, the calcium channel agonist Bay K8644, and calcium-mobilizing thyrotropin-releasing hormone, without affecting accompanying calcium signaling. GSK-A1, a specific inhibitor of PI4KA, also inhibited calcium-driven PRL secretion without affecting calcium signaling or Prl gene expression. In contrast, PIK93, a specific inhibitor of PI4KB, and ISA2011B and UNC3230, specific inhibitors of PIP5K1A and PIP5K1C, respectively, did not affect PRL release (Figure 1). These experiments revealed a key role of PI4KA in calcium-secretion coupling in pituitary lactotrophs downstream of voltage-gated and PI(4,5)P2–dependent calcium signaling [Reference 1].
Figure 1.
PI4-kinase controls basal and receptor-stimulated exocytosis in pituitary lactotrophs independently of PI(4,5)P2.
(a–c) Inhibition of basal prolactin (PRL) release by wortmannin (Wm) and GSK-A1 in perfused (a) and static pituitary cells (b) without affecting de novo PRL synthesis (c).
(d–f) GSK-A1 does not inhibit thyrotropin-releasing hormone (TRH)–stimulated calcium signaling in pituitary lactotrophs (d vs e) but inhibits basal and TRH–stimulated PRL release in perfused pituitary cells (f).
(g–h) GSK-A1 also does not affect stimulated voltage-gated calcium influx by the L-type calcium-channel agonist BayK 8644 (BayK) in pituitary lactotrophs (g vs h) but inhibits BayK–stimulated PRL release in perfused pituitary cells.
Pituitary gonadotroph-specific patterns of gene expression and hormone secretion
Contrary to the hypothesis of pituitary cell plasticity published by several groups, gonadotrophs appear as a single cluster of homogeneous cells, uniquely expressing Fshb, Lhb, and Gnrhr. These cells also specifically express other genes, including Chrna4, Cnga1, Dmp1, Dusp15, Icam5, Lama1, Nhlh2, Nr5a1, Pitx3, Spp1, Tgfbr3l, and Vash2. Some of the genes are clearly expressed in a sex-specific manner, like Dmp1 and its sister gene Spp1. In general, the specific roles of these genes in gonadotroph functions have not yet been elucidated. Our recent studies point to the gonadotroph-specific patterns of gene expression and hormone secretion. The luteinizing hormone (LH) secretory profiles appear to reflect depletion of prestored LH in secretory vesicles by regulated exocytosis. In contrast, follicle-stimulating hormone (FSH) is predominantly released by constitutive exocytosis, and secretory activity reflects the kinetics of Fshb gene expression controlled by gonadotropin-releasing hormone (GnRH), activin, and inhibin. Consistent with the role of activin and inhibin on Fshb expression, pituitary cells express three inhibin subunit genes: Inha is expressed in all hormone-producing cell types and folliculostellate cells (FSCs) in the anterior pituitary and pituicytes in the posterior pituitary; Inhba is expressed only in FSCs and pituicytes, and Inhbb is expressed in gonadotrophs, corticotrophs, FSCs, pituicytes, and pituitary endothelial cells [Reference 2].
Our ongoing work in this project focuses on the roles of the neuroendocrine marker genes Ptprn and Ptprn2, which encode the protein tyrosine phosphatase receptors N and N2, on pituitary gonadotroph function. To do this, we analyzed the effects of their double knockout (DKO) in mice on the hypothalamic-pituitary-gonadal axis. In DKO females, delayed puberty and lack of ovulation were observed, complemented by changes in ovarian gene expression and steroidogenesis. In contrast, testicular gene expression, steroidogenesis, and reproductive organ development were not significantly affected in DKO males. However, in both sexes, pituitary Lhb gene expression and LH levels were reduced, as well as the Fshb gene, while the calcium-mobilizing and LH secretory actions of GnRH were preserved. Hypothalamic Gnrh1 and Kiss1 gene expression was also reduced in DKO females and males. In parallel, a significant reduction in the density of immunoreactive GnRH and kisspeptin fibers was detected in the hypothalamic arcuate nucleus of DKO females and males, while kisspeptin immunoreactivity in the rostral periventricular region of the third ventricle was lowered only in DKO females. These experiments in progress indicate a critical role of Ptprn and Ptprn2 in kisspeptin–GnRH neuronal function and sexual dimorphism in the threshold levels of GnRH required to preserve reproductive function.
A transcriptomics perspective of pituitary corticotrophs
Our scRNA-Seq studies using rat pituitary cells revealed that the corticotroph transcriptome profile was most comparable to that of melanotrophs, and generally agrees with previous basic and clinical work with these cells. However, data on pituitary scRNA-Seq from several species indicate homogeneity of postnatal corticotrophs, forming a distinct cluster of cells, compared with melanotrophs, other hormone-producing cells, and non-hormonal pituitary cells. Genes specific for corticotrophs include Clrn1, Chrna1, Adh1, Angptl8, Hspb3, Lmx1a, Scube2, and Trdn. The roles of these genes in corticotroph functions have not been characterized. Certainly, the most critical genes for corticotroph functions are Crhr1 and Avpr1, encoding the G protein–coupled receptors CRHR1 and AVPR1b, activated by hypothalamic corticotropin-releasing hormone (CRH) and arginine-vasopressin (AVP), respectively. Avpr1b is specifically expressed in corticotrophs, whereas Crhr1 has been detected in some melanotrophs as well. Other genes are specific for melanotrophs, including Oacyl, Pax7, Esm1, and Pcsk2. Moreover, scRNA-Seq data provide a wealth of new information regarding the expression of several common genes encoding other G protein–coupled receptors and enzyme-linked plasma membrane receptors, and their signal transduction pathways, which have not previously been reported to be expressed in the pituitary gland. The expression pattern of these receptors and their ligands highlight the importance of autocrine/paracrine regulation of pituitary cell function and the modulating role of peripheral glands through nuclear receptors [Reference 3].
We also contributed to the work of our collaborator Prashant Chittiboina on scRNA-Seq studies of human hormone-producing pituitary adenomas causing Cushing’s disease (CD). The analysis included over 25,000 cells and identified unique CD adenoma transcriptomic signatures compared with adjacent normal cells, with validation by bulk RNA-Seq, DNA methylation, qRT-PCR, and immunohistochemistry. CD adenoma cells include a subpopulation of proliferating, terminally differentiated corticotrophs. In CD adenomas, we found recurrent promoter hypomethylation and transcriptional upregulation of PMAIP1 (encoding proapoptotic BH3-only bcl-2 protein noxa) but paradoxical noxa downregulation. Using primary CD adenoma cell cultures and a corticotroph-enriched mouse cell line, we found that selective proteasomal inhibition with bortezomib stabilizes noxa and induces apoptosis, indicating its utility as an anti-tumor agent [Reference 4].
The astroglial and stem cell functions of adult pituitary folliculostellate cells
Recently, we presented scRNA-Seq and immunohistofluorescence analyses of pituitary cells of adult female rats, with a focus on the transcriptomic profiles of nonhormonal cell types. Samples obtained from whole pituitaries and separated anterior and posterior lobe cells contained all expected pituitary-resident cell types and lobe-specific vascular cell subpopulations. FSCs and pituicytes expressed S100B, ALDOC, EAAT1, ALDH1A1, and VIM genes and proteins, as well as other astroglial marker genes, some common and some cell-type specific. We also found that the SOX2 gene and protein were expressed in about 15% of pituitary cells, including FSCs, pituicytes, and a fraction of hormone-producing cells, arguing against its stem-cell specificity. FSCs comprised two Sox2-expressing subclusters; one larger (FS1, 1,877 cells) and one smaller (FS2, 511 cells). Interestingly, FS2 cells expressed a much greater diversity of genes than did FS1 cells, with a median of 3,137 and 2,099 genes detected per cell, respectively. The finding that 80% of FS1–upregulated genes are also upregulated in FS2 suggests that FS1 and FS2 cells are closely related cells. In contrast, only 30% of FS2–upregulated genes were pan–FSC genes, suggesting that these cells have additional activated gene-expression programs. We also noticed that FSCs shared expression of several genes with hormone-producing cells. However, a majority of these were dominantly expressed by FS2 compared with FS1 (Figure 2). We also found that FS1 cells were randomly distributed in the anterior and intermediate lobes, while FS2 cells were localized exclusively in cells of the marginal zone between the anterior and intermediate lobes. These data indicate that the identity of the FSCs are specialized anterior pituitary–specific astroglia, with FS1 cells representing differentiated cells, with transcriptomes consistent with classical FSC roles, and FS2 cells exhibiting additional stem cell-like features [Reference 5].
Figure 2. Identification of two FSC subtypes
(a) UMAP embedding showing unsupervised clustering of FSCs into two subtypes. A small portion of FS2 cells (circled) express cell cycle markers.
(b) Violin plot of the distributions of number of genes per cell in FS1 and FS2, with median values of 2,099 and 3,137, respectively.
(c) Venn diagram showing the number of genes identified as FS1–dominant, FS2–dominant, and common to both, relative to both cell types except pituicytes.
(d) Dot plot showing selected top FSC–expressed genes: dominant in both subtypes (first group), FS1–dominant (second group), FS2–dominant (third group), and co-expressed in FS2 and HPC (fourth group).
(e) UMAP scatter plots showing the expression patterns of selected genes from panel d.
Publications
- Kucka M, Gonzales-Iglesisas A, Tomic M, Previde RM, Smiljanic K, Sokanovic SJ, Fletcher PA, Sherman A, Balla T, Stojilkovic SS. Calcium-prolactin secretion coupling in female rat pituitary lactotrophs is controlled by PI4-kinase alpha. Front Endocrinol 2021 12:790441.
- Constantin S, Bjelobaba I, Stojilkovic SS. Pituitary gonadotroph-specific patterns of gene expression and hormone secretion. Curr Opin Pharmacol 2022 66:102274.
- Stojilkovic SS, Previde RM, Sherman AS, Fletcher PA. Pituitary corticotroph identity and receptor-mediated signaling: A transcriptomics perspective. Curr Opin Endocr Metab Res 2022 25:100364.
- Asuzu DT, Alvarez R, Fletcher PA, Mandal D, Johnson K, Wu W, Elkahloun A, Clavijo P, Allen C, Maric D, Ray-Chaudhury A, Rajan S, Abdullaev Z, Nwokoye D, Aldape K, Nieman LK, Stratakis C, Stojilkovic SS, Chittiboina P. Pituitary adenomas evade apoptosis via noxa deregulation in Cushing's disease. Cell Rep 2022 40:111223.
- Fletcher PA, Smiljanic K, Prévide RM, Constantin S, Sherman AS, Coon SL, Stojilkovic SS. The astroglial and stem cell functions of adult rat folliculostellate cells. Glia 2022 10:1002.
Collaborators
- Tamás Balla, MD, PhD, Section on Molecular Signal Transduction, NICHD, Bethesda, MD
- Prashant Chittiboina, MD, PhD, Neurosurgery Unit for Pituitary and Inheritable Diseases, NINDS, Bethesda, MD
- Patrick A. Fletcher, PhD, Laboratory of Biological Modeling, NIDDK, Bethesda, MD
- Arthur Sherman, PhD, Laboratory of Biological Modeling, NIDDK, Bethesda, MD
Contact
For more information, email stojilks@mail.nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/stojilkovic.