Cell Cycle Regulation in Oogenesis

- Mary Lilly, PhD, Head, Section on Gamete Development
- Chun-yuan Ting, PhD, Staff Scientist
- Kuikwon Kim, MS, Biologist
- Shu Yang, PhD, Postdoctoral Fellow
- Lucia Bettedi, PhD, Visiting Fellow
- Yingbiao Zhang Zhang, PhD, Visiting Fellow
Our long-term goal is to obtain a comprehensive understanding of how metabolic signaling pathways influence oocyte growth, development, and quality. Chromosome mis-segregation during female meiosis is the leading cause of miscarriages and birth defects in humans. Recent evidence suggests that many meiotic errors occur downstream of defects in oocyte growth and/or in the hormonal signaling pathways that drive differentiation of the oocyte. Thus, understanding how oocyte development and growth impact meiotic progression is essential to studies in both reproductive biology and medicine. We use the genetically tractable model organism Drosophila melanogaster to examine how meiotic progression is instructed by the developmental and metabolic program of the egg.
In mammals, studies on the early stages of oogenesis face serious technical challenges in that entry into the meiotic cycle, meiotic recombination, and the initiation of the highly conserved prophase I arrest all occur during embryogenesis. By contrast, in Drosophila these critical events of early oogenesis all take place continuously within the adult female. Easy access to the early stages of oogenesis, coupled with available genetic and molecular-genetic tools, makes Drosophila an excellent model for studies on the role of metabolism in oocyte development and maintenance.
The GATOR complex: integrating developmental and metabolic signals in oogenesis
The Target of Rapamycin Complex 1 (TORC1) regulates cell growth and metabolism in response to many inputs, including amino-acid availability and intracellular energy status. In the presence of sufficient nutrients and appropriate growth signals, the Ragulator and the Rag GTPases (a complex that regulates lysosomal signaling and trafficking) target TORC1 to lysosomal membranes, where TORC1 associates with its activator, the small GTPase Rheb. Once activated, TORC1 is competent to phosphorylate its downstream targets. The Gap activity towards Rags (GATOR) complex is an upstream regulator of TORC1 activity.
The GATOR complex consists of two subcomplexes (Figure 1). The GATOR1 complex inhibits TORC1 activity in response to amino-acid starvation. GATOR1 is a trimeric protein complex consisting of the proteins Nprl2, Nprl3, and Iml1. Evidence from yeast and mammals indicates that the components of the GATOR1 complex function as GTPase–activating proteins (GAP) that inhibit TORC1 activity by inactivating the Rag GTPases. Notably, Nprl2 and Iml1 are tumor-suppressor genes, while mutations in Iml1, known as DEPDC5 in mammals, are a leading cause of hereditary epilepsy.
Figure 1. The GATOR complex regulates TORC1 activity.
The GATOR2 complex opposes the activity of the TORC1 inhibitor GATOR1.
Our work, as well as that of others, found that the GATOR2 complex activates TORC1 by opposing the TORC1–inhibitory activity of GATOR1. Intriguingly, computational analysis indicates that Mio and Seh1, as well as several other members of the GATOR2 complex, have structural features consistent with coatomer proteins and membrane-tethering complexes. In line with the structural similarity to proteins that influence membrane curvature, we showed that three components of the GATOR2 complex, Mio, Seh1, and Wdr24, localize to the outer surface of lysosomes, the site of TORC1 regulation. However, how GATOR2 inhibits GATOR1 activity, thus allowing for the robust activation of TORC1, remains unknown. Additionally, the role of the GATOR1 and GATOR2 complexes in both the development and physiology of multicellular animals remains poorly defined. Over the past year, we used molecular, genetic, and cell-biological approaches to define the in vivo functions of the GATOR1 and GATOR2 complexes in the model organism Drosophila.
The GATOR2 component Wdr59 promotes or inhibits TORC1 activity depending on cellular context.
As mentioned above, the GATOR2 complex inhibits GATOR1 and thus serves to activate TORC1. However, the mechanism by which GATOR2 inhibits GATOR1 remains unknown. In its initial functional characterization in mammalian and Drosophila cultured cells, the GATOR2 complex was reported to contain five protein Mios/Mio, Seh1, Sec13, Wdr24, and Wdr59 (Figure 1A). In these earlier studies, knockdowns of GATOR2 components resulted in constitutive activation of GATOR1 and reduced TORC1 activity. Similarly, Drosophila mutants of the GATOR2 components mio, seh1, and wdr24 exhibit reduced TORC1 activity and growth in the female germline. However, a recent study from Schizosaccharomyces pombe reported that SEA3/WDR59 inhibits TORC1 activity as a component of the GATOR1 complex. Notably, this is the opposite of the role assigned to Wdr59 based on studies in both human and Drosophila cultured cells. Additionally, deletions of Wdr59 in HEK293 (human embryonic kidney) cells and mouse embryonic fibroblasts result in a partial resistance to nutrient deprivation. Thus, the exact function of Wdr59 within the GATOR–TORC1 signaling pathway remains unclear.
Over the last year, we showed that Wdr59, originally assigned to the GATOR2 complex based on studies performed in tissue culture cells, unexpectedly has a dual function in TORC1 regulation in Drosophila. We demonstrated that, in the ovary and the eye imaginal disc brain complex, Wdr59 inhibits TORC1 activity by opposing the GATOR2–dependent inhibition of GATOR1. Conversely, in the Drosophila fat body, Wdr59 promotes the accumulation of the GATOR2 component Mio and is required for TORC1 activation. Similarly, in mammalian HeLa cells, Wdr59 prevents the proteolytic destruction of GATOR2 proteins Mio and Wdr24. Consistent with the reduced levels of the TORC1–activating GATOR2 complex, Wdr59 knockout (KO) HeLa cells have reduced TORC1 activity, which is restored along with GATOR2 protein levels upon proteasome inhibition. Taken together, our data support a model in which the Wdr59 component of the GATOR2 complex functions to promote or inhibit TORC1 activity depending on cellular context. These studies broaden our understanding of the GATOR–TORC1 signaling axis in metazoans and highlight the complexity of metabolic regulation in metazoans.
Many independent pathways converge on Nprl3 to regulate TORC1 activity in Drosophila.
In collaboration with the laboratory of Youheng Wei, we characterized several pathways that regulate the expression of the GATOR1 component Nprl3 in Drosophila (Figure 2). We determined that the stability of Nprl3 is impacted by the Unassembled Soluble Complex Proteins Degradation (USPD) pathway. In addition, we found that FK506 binding protein 39 (FKBP39)–dependent proteolytic destruction maintains Nprl3 at low levels in nutrient-replete conditions. Nutrient starvation abrogates the degradation of the Nprl3 protein and rapidly promotes Nprl3 accumulation. Consistent with a role in promoting the stability of a TORC1 inhibitor, mutations in fkbp39 reduced TORC1 activity and increased autophagy. We also demonstrated that the 5′ UTR of nprl3 transcripts contain a functional upstream open-reading frame (uORF) that inhibits main ORF translation. In summary, our work uncovered novel mechanisms of Nprl3 regulation and identified an important role for Drosophila FKBP39 in the control of cellular metabolism and growth.
Figure 2. Many pathways regulate the levels of the TORC1 inhibitor Nprl3.
A. The nprl3 mRNA contains a functional uORF that reduces Nprl3 translation.
B. Nprl3 forms the trimeric GATOR1 complex with the proteins Nprl2 and Iml1. When not assembled into the GATOR1 complex, Nprl3 is degraded via a pathway dependent on the ubiquitin ligase HUWE1.
C. In nutrient-replete conditions, FKBP39 associates with Nprl3 and promotes its degradation. Upon amino acid starvation, the FKBP39–dependent destruction of Nprl3 is blocked, and the increased levels of GATOR1 result in reduced TORC1 activity.
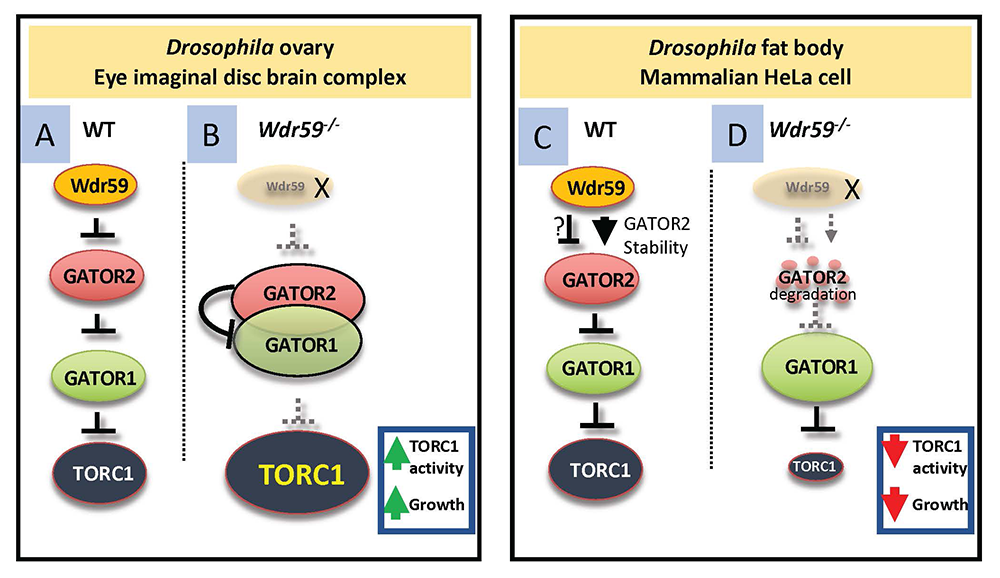
Figure 3. Integrated model of TORC1 regulation
A. In wild-type animals, Wdr59 inhibits TORC1 activity upstream of the GATOR2 complex in the ovary and eye imaginal disc brain complex.
B. In wdr59 mutants, GATOR2 increases its association with GATOR1, further inhibiting GATOR1 activity, and allowing for the increased activation of TORC1.
C. In the fat body of Drosophila and mammalian HeLa cells, Wdr59 protects GATOR2 components from degradation by the proteasome.
D. In the absence of Wdr59 from the Drosophila fat body or from HeLa cells, GATOR2 components are destroyed by the proteasome, resulting in derepression of GATOR1, increased interaction of GATOR1 with RagA, and concomitant reduction in TORC1 activity. Note that the GATOR1 complex regulates TORC1 activity by acting as a GAP for the RagA component of the GTPase, which functions to activate TORC1.
Publications
- Zhou Y, Guo J, Wang X, Cheng Y, Guan J, Barman P, Sun MA, Fu Y, Wei W, Feng C, Lilly MA, Wei Y. FKBP39 controls nutrient dependent Nprl3 expression and TORC1 activity in Drosophila. Cell Death Dis 2021;12(6):571.
Collaborators
- Brian Calvi, PhD, Indiana University, Bloomington, IN
- Ryan Dale, PhD, Bioinformatics and Scientific Programming Core, NICHD, Bethesda, MD
- Thomas Gonatopoulos-Pournatzis, PhD, Functional Transciptomics Section, NCI, Frederick, MD
- Mark Nellist, PhD, Erasmus University Medical Center, Rotterdam, Netherlands
- Youheng Wei, PhD, Yangzhou University, Yangzhou, Jiangsu, China
Contact
For more information, email mlilly@helix.nih.gov or visit https://irp.nih.gov/pi/mary-lilly.