Mechanisms of Nuclear Genome Organization and Maintenance

- Mary Dasso, PhD, Head, Section on Cell Cycle Regulation
- Vasilisa Aksenova, PhD, Contract Research Scientist
- Alexei Arnaoutov, PhD, Staff Scientist
- Maia Ouspenskaia, DVM, Biologist
- Abik Nandi, PhD, Visiting Postdoctoral Fellow
- Sanjana Sundararajan, PhD, Visiting Postdoctoral Fellow
- Reham Aljumaah, BA, Predoctoral Intramural Research Training Award Fellow
- Sierra Ball, BA, Postbaccalaureate Intramural Research Training Award Fellow
- Carolyn Egekeze, BA, Postbaccalaureate Intramural Research Training Award Fellow
- Elizabeth Giordano, BA, Postbaccalaureate Intramural Research Training Award Fellow
- Jacob Miller, BA, Postbaccalaureate Intramural Research Training Award Fellow
- Ashley Person, BA, Postbaccalaureate Intramural Research Training Award Fellow
We are interested in mechanisms of genome maintenance and organization. During interphase, chromosomes are surrounded by the nuclear envelope (NE), which separates the nuclear from cytoplasmic compartment of the cell. The sequestration of chromosomes within the nucleus has profound consequences for almost all aspects of gene expression and cell function. Communication between the nucleus and cytoplasm occurs through conduits called nuclear pore complexes (NPCs), which are embedded in the NE and consist of about 34 proteins called nucleoporins (Figure 1). Beyond nucleo-cytoplasmic trafficking, nucleoporins are important for chromosome organization, transcriptional control, RNA processing, cell signaling, and cell-cycle control. Both nucleoporins and soluble components of the nuclear trafficking machinery also perform transport-independent functions in mitotic chromosome segregation. The involvement of nucleoporins in such diverse events offers the intriguing possibility that they might coordinate these processes with nuclear trafficking and with each other. Moreover, nucleoporin dysfunction has important clinical implications: nucleoporin genes are frequently misregulated in cancers, and nucleoporin mutations cause congenital defects, pediatric nephrotic syndromes, and premature ovarian insufficiency. Nucleoporins are critical viral targets, and their disruption contributes to neurodegenerative conditions, including amyotrophic lateral sclerosis, frontotemporal dementia, and Huntington’s disease.
Our research centers on nucleoporins, NPC–associated proteins (e.g., the SUMO pathway, spindle-checkpoint proteins), and other components of the nuclear transport machinery (e.g., the Ran pathway) throughout the cell cycle. Our goal is to define their biochemical roles and how their dysregulation causes human disease. We took a multifaceted approach toward this question, using both CRISPR–based strategies in mammalian cells and fly (Drosophila melanogaster) developmental genetics.
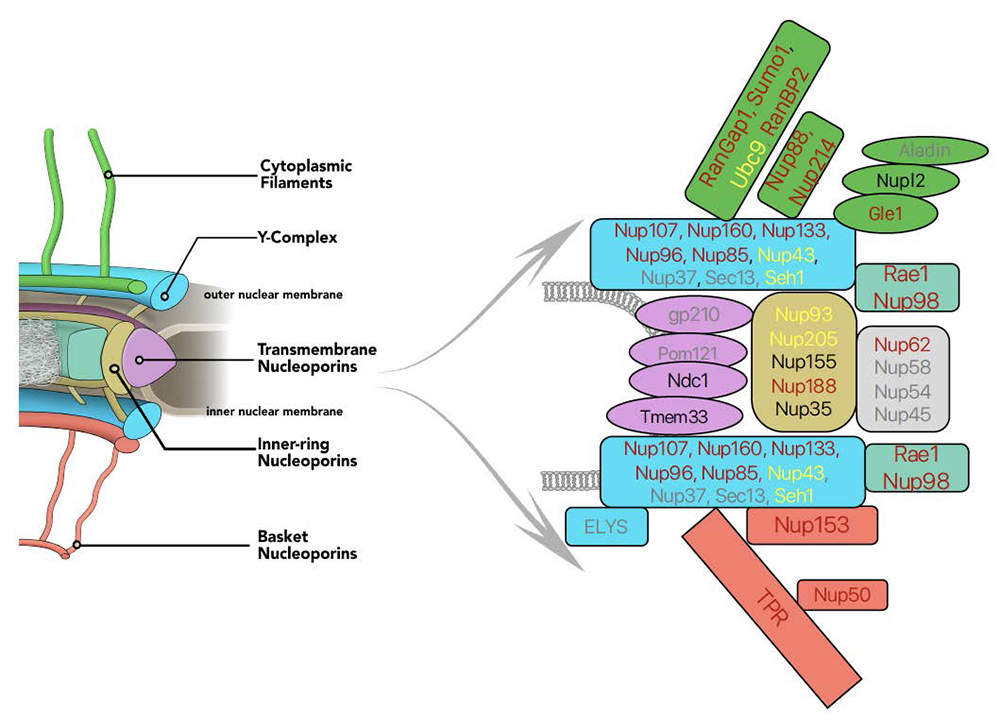
Figure 1. Auxin-induced degron (AID) tagging of nucleoporins
Left. Model of nuclear pore structure with cytoplasmic filaments (green) oriented upward and nucleoplasmic basket (reddish brown) oriented downward. Other domains of the NPC as indicated.
Right. Polypeptides associated with NPC sub-complexes, with fill colors corresponding to model domains. Text shows progress in obtaining AID–tagged cell lines for individual nucleoporins: reddish brown: successful tagging; black: in progress; yellow: tagging achieved but cell-line quality issues; grey: tagging unsuccessfully attempted.
Selective degradation of nucleo-cytoplasmic transport proteins and their interphase function
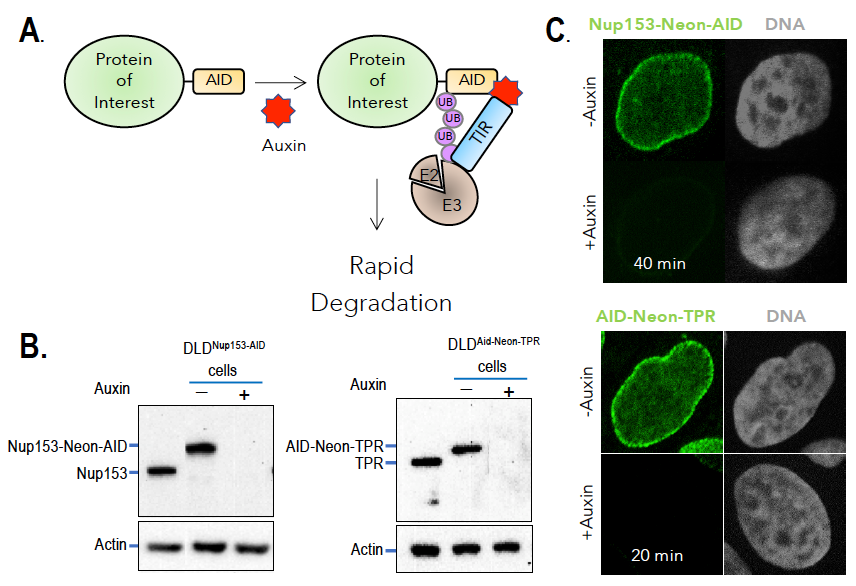
Understanding the activities of individual nucleoporins has been complicated by their multifaceted nature, abundance, and unusual stability. To overcome these issues, we employed strategies for selective and rapid degradation of individual proteins within human tissue-culture cells (Figure 2). Specifically, we used CRISPR-Cas9 to construct cell lines in which sequences encoding auxin-induced degron (AID) domains are inserted into both alleles of targeted genes within cells that also stably express the transport inhibitor response 1 (TIR1) protein. TIR1 promotes rapid, selective degradation upon addition of the plant hormone auxin. Frequently, we also add a fluorescent tag to the targeted proteins, allowing their degradation to be monitored visually as well as biochemically. We have been successful in developing cell lines that allow conditional depletions of nucleoporins associated with different regions of the NPC (Figure 1).
Our recent findings regarding the roles of nucleoporins during interphase address three issues. First, we investigated the role of individual nucleoporins in NPC assembly and stability. Our results indicate that different regions of the NPC can persist independently after depletion of individual nucleoporins, suggesting that the NPC is a surprisingly modular structure. Second, we investigated the role of individual nucleoporins in different nuclear trafficking pathways, an assessment that includes evaluation of nuclear protein import, protein export, and RNA export. We are able to differentiate between the roles of individual nucleoporins and can specifically show that the TPR protein has a unique and important role in nuclear mRNA export via the transcription-export-2 (TREX-2) complex (see also below). Third, we investigated how organization of the nuclear transport machinery impacts development. In particular, we found that an evolutionarily controlled mechanism for association of Ran’s GTPase–activating protein (RanGAP) is critical for the development of Drosophila. Defining the mechanism through which individual nucleoporins contribute to each of these processes will allow us to better design future experiments examining nucleoporin function in human development and disease.
Figure 2. Auxin-induced degradation of AID–tagged nucleoporins
A. TIR1–expressing cells ubiquitinate and degrade proteins tagged with AID domains upon auxin addition. Nup153 and TPR were homozygously fused with the AID tag and a fluorescent marker (neon green) in TIR1–expressing cells. Rapid and uniform degradation is observed after auxin addition by Western blotting (B) or destruction of the fluorescent tag (C).
Analysis of nucleoporins demonstrates that the NPC is a highly modular structure.
NPCs are built from many copies of roughly 34 distinct nucleoporins. Models of the NPC depict it as a composite of several sub-domains, which have been named the outer rings, inner ring, cytoplasmic fibrils, and nuclear basket. The outer-ring domains of the NPC are formed from the Y-complex, which contains nine core nucleoporins (SEH1, SEC13, NUP37, NUP43, NUP85, NUP96, NUP107, NUP133, and NUP160), with a tenth subunit (ELYS) required for chromatin recruitment. Other nucleoporins (NUP205, NUP188, NUP155, NUP93, and NUP35) form the inner-ring structures. The distinct roles of individual nucleoporins and their functional interactions remain poorly understood. Moreover, NPCs undergo a disassembly-reassembly cycle during mitotic division, and a lack of tools for acute manipulation of individual nucleoporins has therefore precluded the study of their roles in maintaining structures within pre-existing pores without complications from disruption of NPC assembly.
We added AID tags and fluorescent moieties by homozygously targeting gene loci encoding Y-complex and inner-ring nucleoporins. Auxin addition resulted in a rapid loss of the targeted proteins in each case, without degradation of other nucleoporins. We anticipated that loss of any Y-complex member should result in complete destabilization of the outer rings. While this was true after depletion of NUP96 or NUP107, the loss of other Y-complex members surprisingly left the outer-ring lattice in place. The findings suggest that the outer-ring structure is remarkably resistant to perturbations, once it is fully assembled, and show that its members are not of equivalent importance in sustaining its stability. Importantly, near-complete loss of the outer ring in NUP96–depleted cells did not cause collapse of the rest of the NPC, as demonstrated by immunostaining, live microscopy, and mass spectrometry. Particularly, the remarkable persistence of inner-ring nucleoporins indicated resilience of the NPC structure. Interestingly, depletion of the inner-ring nucleoporin NUP188 caused an NPC disassembly that was opposite to the profile after NUP96 depletion: inner-ring components were extensively displaced, while the components of the cytosolic fibrils, outer ring, and basket were largely unaffected. Also, there was a global reduction of almost all nucleoporins upon loss of NUP93. High-resolution scanning electron microscopy of residual NPCs after depletion of NUP96 or NUP93 further confirmed the status of these structures. Together, our results indicate that the inner and outer rings of the NPC form distinct and independent structures, and that NUP93 serves as an NPC lynchpin essential for both of them (Figure 3).
Figure 3.
Schematic representations of NPC structure upon loss of NUP188, NUP96, or NUP93
After depletion of the inner ring or outer rings, we tested whether the residual structures remained functional for the import and export of a model nuclear transport substrate. Remarkably, there were only minimal changes in both nuclear import and export rates upon loss of NUP96 or NUP188. However, NUP93 depletion caused a complete block in nuclear transport in both directions, confirming that global disruption functionally disabled NPCs. Taken together, the results indicate that persistent inner-ring or outer-ring structures could still act as conduits for vectoral nuclear trafficking and that these modules can support independent and redundant trafficking routes. Removal of both sets of structures forecloses all nuclear trafficking. Notably, the persistence of functional pores lacking a subset of canonical nucleoporins suggests that terminally differentiated cells might retain substantial nuclear trafficking even with divergent NPC composition. Differentiated cells might thus customize function through altered NPC composition, potentially modulating specific trafficking pathways or aspects of NPC activity, such as gene regulation and post-translational protein modifications.
Roles of nucleoporins in gene expression and RNA trafficking
A series of evolutionarily conserved complexes are co-transcriptionally recruited to nascent mRNAs, facilitating their processing as well as escorting them to and through the NPC, actions that are functionally linked; a failure to perform any of them during mRNA biogenesis directly impacts both upstream and downstream events. A key player in mRNA maturation is the transcription and export 2 (TREX-2) complex. Loss of the TREX-2 complex leads to defects in mRNA export, similar to the phenotype observed after loss of the major mRNA export receptor NXF1. The GANP subunit of TREX-2 localizes within the nucleus and associates with the NPC's nuclear basket, which protrudes from the nucleoplasmic face of the NPC. In vertebrates, the nuclear basket comprises three nucleoporins (BSK-NUPs), called NUP153, TPR, and NUP50. BSK-NUPs have been implicated in numerous processes beyond protein import and export, including chromatin remodeling, control of gene expression, and protein modification, as well as mRNA processing and export.
It has been difficult to analyze discrete NPC functions in the absence of vertebrate BSK-NUPs; knockout of their genes is deleterious for organisms, and their depletion by RNAi requires extended incubations, potentially permitting the emergence of secondary phenotypes from prolonged NPC disruption or defective post-mitotic NPC re-assembly. We used AID to untangle the functions of individual BSK-NUPs in both nuclear-basket architecture and gene expression. We found that NUP153 and TPR bound to the NPC independently of each other and that loss of individual BSK-NUPs did not destabilize the NPC. We further found that TPR, but not NUP153 or NUP50, tethers the TREX-2 complex to the NPC. Loss of NUP50, NUP153, or TPR led to unique transcriptomic responses. Importantly, transcriptomic signatures after loss of TPR were more pronounced and similar to changes upon the loss of either the GANP subunit of the TREX-2 complex or of the RNA–export receptor NXF1. Moreover, similar to the case of NXF1 or GANP, loss of TPR led to retention of both upregulated and downregulated mRNA transcripts within the nucleus.
Taken together, these data support a unique role of TPR in transcription regulation and mRNA export through the TREX-2 complex. The findings both support our hypothesis that individual nucleoporins have distinct and non-redundant cellular functions, and they demonstrate the utility of the AID system to analyze their unique roles within cells.
Impact of Ran pathway organization beyond nuclear trafficking: development and mitosis
The Ran GTPase is a critical regulator of nuclear trafficking in eukaryotic cells. Ran’s cytoplasmic GTPase–activating protein, RanGAP, and its chromatin-bound nucleotide exchange factor, RCC1, establish compartmental patterns for Ran’s nucleotide-binding status: GTP–bound Ran (Ran-GTP) is abundant in the nucleus, while GDP–bound Ran (RanGDP) is abundant in the cytosol. The asymmetry drives nucleo-cytoplasmic trafficking through controlled cargo binding and release by karyopherins, a family of RanGTP–binding transport receptors.
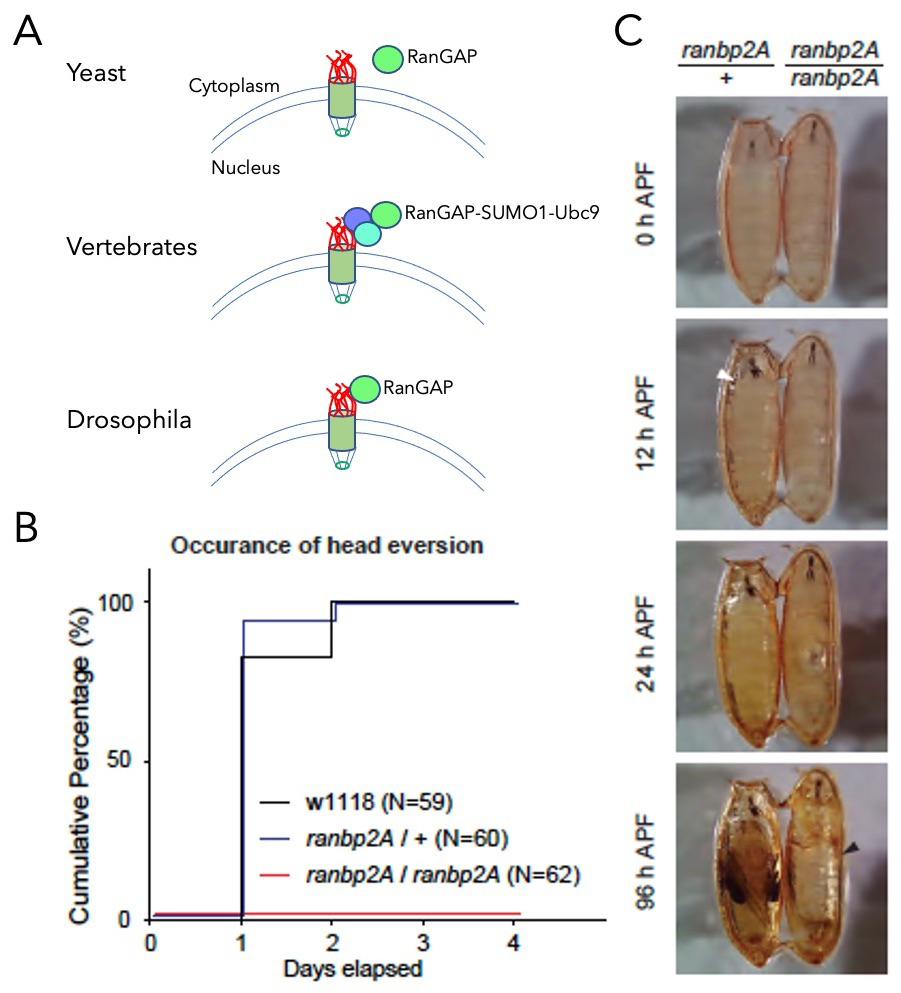
In multicellular organisms, RanGAP is tethered to the cytoplasmic side of the NE, but the functional consequences of its localization had not been clarified. To investigate the importance of RanGAP association with the NE, we used human tissue-culture cells and Drosophila. In mammals, RanGAP tethering is mediated by its covalent conjugation to SUMO1, a small ubiquitin-like protein. We made human tissue-culture cells in which RanGAP was neither SUMOylated nor localized to the NE. Surprisingly, the cells showed neither obvious changes in viability nor substantial defects in nucleo-cytoplasmic transport of a model substrate. In Drosophila, we found that SUMOylation of RanGAP did not control its association with the NE. Rather, we identified a specific region within the nucleoporin dmRanBP2 (Drosophila melanogaster RanBP2) that directly binds to dmRanGAP, tethering it to the NPC (Figure 4). A dmRanBP2 mutant lacking this region showed no apparent growth defects during larval stages, but arrested at the early pupal stage. The developmental arrest was rescued by a direct fusion of dmRanGAP to the dmRanBP2 mutant, indicating that recruitment of dmRanGAP to dmRanBP2 per se was necessary for the pupal ecdysis sequence during development. Collectively, the results indicate that, while the localization of dmRanGAP to the NE is widely conserved in multicellular organisms, the targeting mechanisms are not. Furthermore, the localization appears to be critical for developmental processes rather than for viability at the cellular level. We continue to explore the developmental consequences of releasing the interaction of dmRanGAP to dmRanBP2.
Figure 4. Anchorage of dmRanGAP to dmRanBP2 is required for development.
A. Schematic representation of RanGAP association to NPCs. In yeast, RanGAP (green) is not bound to the NPC (top). In vertebrates, RanGAP binds to RanBP2 (red) in a complex that also contains Ubc9 (aqua) and SUMO1 (blue, middle). In flies, dmRanGAP binds directly to dmRanBP2 (bottom).
B. Cumulative step histogram of head eversion event during pupal stage. Late 3rd instar larvae (5–day old) were collected on day 0. While head eversion occurred robustly in controls (both w1118 and heterozygous ranbp2A flies), no head eversion event was observed in homozygous ranbp2A flies.
C. Behavior analysis of pupal ecdysis. Heterozygous or homozygous ranbp2A flies were collected at the white pupal stage (APF: after puparium formation). The homozygous ranbp2A fly failed to undergo head eversion (white arrowhead) and air bubble translocation after 24 hours, and the air bubble occupied most of the pupa (black arrowhead) after 96 hours.
We have a long-standing interest in the process of chromosome segregation. The Ran GTPase pathway and nucleoporins promote chromosome segregation through their important roles in spindle assembly and cell-cycle progression. After mitotic NE breakdown, RCC1 generates RanGTP near chromosomes, while Ran distal to chromosomes is GDP–bound, directing spindle assembly through spatially regulated release of spindle assembly factors (SAFs) from karyopherins. To segregate chromosomes accurately, RanGTP distribution must be tightly regulated, both spatially and temporally. Defects in chromosome segregation lead to aneuploidy, a condition in which cells possess an abnormal number of chromosomes. The error-prone nature of Ran–dependent spindle assembly is particularly important during meiosis, potentially contributing to human pregnancy losses and genetic disorders, including Down’s syndrome. Moreover, aneuploidy arising from mitotic divisions is a hallmark of many solid tumors.
RanBP1 is a RanGTP–binding protein that forms a stable heterotrimeric complex with Ran and RCC1 (RRR complex) in vitro, inhibiting RCC1’s nucleotide exchange activity. We previously reported that RRR complex formation determines RCC1’s partitioning between its chromatin-bound and soluble forms in embryonic systems, where RCC1 levels are very high, as well as specifically inhibiting the activity of soluble RCC1. Somatic cells have less soluble RCC1 during mitosis, raising questions about whether RRR complex formation is important after early development. To investigate the mitotic role of the RRR complex after early development, we examined whether RanBP1 or a related protein called RanBP3 might be important for controlling mitotic RanGTP gradients within somatic cells. We systematically varied RanBP1 and RanBP3 levels in HCT116 or DLD1 cell lines through overexpression or fusion with AID tags. Consistent with earlier reports, RanBP1 was dispensable for interphase import or export of a model substrate, while RanBP3 appears to facilitate nuclear export via the Crm1 karyopherin. Within mitosis, altering RanBP1 levels substantially altered RCC1 dynamics on metaphase chromosomes, while altering RanBP3 levels did not. Moreover, we found dramatic re-localization of the SAF hepatoma up-regulated protein (HURP, a component of the spindle-assembly pathway) during metaphase, in direct correspondence with changes in RCC1 dynamics (Figure 5), showing that RanGTP levels and SAF activity near chromosomes correlate with altered RCC1 behavior. Analogous to findings in embryonic systems, the data indicate an important mitotic role in human somatic cells for RanBP1 in controlling RCC1 dynamics and determining the accurate spatial distribution and magnitude of Ran-GTP gradients, thus ensuring correct execution of Ran–dependent mitotic events.
Figure 5. Regulation of mitotic Ran–GTP gradients by the RRR complex
Immuno-staining with HURP and tubulin antibodies of cells in HCT116RCC1-µAID-3xFLAG, HCT116RanGAP1-3mAID, and HCT116RanBP1-µAID-HA cells treated with or without 1 mM Auxin for 5, 2, and 3 h, respectively. Cells also express TIR1. White dashed lines represent the length of HURP signal (top row). Scale bars = 10 μm.
The role of the IRBIT protein in tissue homeostasis
We previously reported a conserved role for the IRBIT protein (IP3-receptor–binding protein released with inositol 1,4,5-trisphosphate) in inhibiting ribonucleotide reductase (RNR), an enzyme that produces deoxynucleotide triphosphates (dNTPs) for DNA synthesis. We further found that mammalian tissue-culture cells show altered cell-cycle progression and genome stability in the absence of IRBIT, and that the mechanism is conserved between humans and Drosophila. Therefore, in collaboration with Mihaela Serpe and Brian Oliver, we used flies as a model organism to understand the role of IRBIT in development and tissue homeostasis.
In situ hybridization showed IRBIT expression in regions destined to become the midgut during embryogenesis, and that IRBIT is highly expressed in the adult midgut. The Drosophila midgut has a tubular structure and is surrounded by visceral muscles. The adult midgut possesses a monolayered epithelium composed of four distinct cell types (Figure 6B): intestinal stem cells (ISCs), undifferentiated progenitor cells called enteroblasts (EBs), specialized absorptive enterocytes (ECs), and secretory enteroendocrine cells (EEs). The midgut is maintained through division of ISCs, giving rise to EBs, which in turn differentiate into EEs. Nutrients are absorbed from the lumen of the gut, which also contains a complex microbiota; the midgut acts both as a niche for commensal microbes and as the first line of defense against microbial pathogens. Like the intestine of vertebrates, the epithelium of the midgut has a remarkable regenerative capacity, which has been extensively exploited for the study of stem cell–driven tissue self-renewal, as well as tissue homeostasis during aging.
We examined IRBIT’s role in the midgut by generating an IRBIT null fly (IRBIT–/–) (Figure 6A). The midguts of one-day-old wild-type and IRBIT–/– flies were essentially indistinguishable at the tissue-architecture level. However, we observed a rapid loss of tissue homeostasis in the IRBIT–/– flies, with a progressive increase in relative numbers of undifferentiated enteroblast progenitor cells and tissue dysplasia. IRBIT–/– flies also show fewer cell-cell contacts, when stained for junctional proteins in the posterior midgut epithelium, and altered gene expression patterns, reminiscent of changes associated with inflammation and aging. The phenotypes are fully rescued by expression of full-length IRBIT; further experiments suggested that altered dNTP pools likely contribute to the IRBIT–/– phenotypes. Further analysis showed that the IRBIT–RNR pathway is essential to ensure correct differentiation of ISC progeny, and that it is a key downstream target of the GATAe transcription factor. Moreover, the GATAe–IRBIT–RNR pathway becomes dysfunctional as flies age, contributing to a characteristic accumulation of undifferentiated ISC progeny, which can be reversed by specifically inhibiting RNR within progeny cells. Collectively, our findings showed that RNR suppression by IRBIT is an important mechanism that directs differentiation of ISC progeny to maintain intestinal tissue homeostasis.
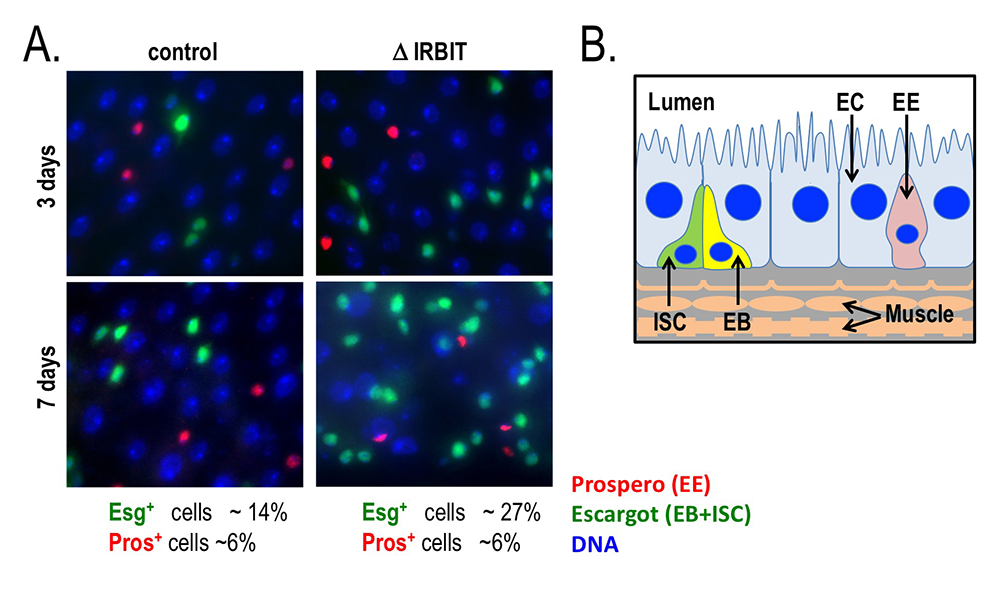
Figure 6. Loss of IRBIT disrupts tissue homeostasis in the Drosophila midgut.
A. Epithelia from control (left) and IRBIT–/– (right) flies at three (top) or seven (bottom) days after eclosure. The guts were stained with the DNA dye Hoechst 33258 (blue), and antibodies against Prospero (red, to identify EE cells) and Escargot (green, to identify EB and ISC cells). We observed a rapid and progressive increase in the fraction of Escargot-positive cells in the IRBIT–/– flies over time. In conjunction with additional experiments, the accumulation is indicative of accumulation of undifferentiated enteroblast progenitor cells.
B. Schematic of epithelium within the Drosophila midgut. EC: enterocyte; ISC: intestinal stem cell; EB: enteroblast; EE: enteroendocrine cell.
Publications
- Li Y, Aksenova V, Tingey M, Yu J, Ma P, Arnaoutov A, Chen S, Dasso M, Yang W. Distinct roles of nuclear basket proteins in directing the passage of mRNA through the nuclear pore. Proc Natl Acad Sci U S A 2021 118:e2015621118.
- Chen S, Lyanguzova M, Kaufhold R, Plevock Haase KM, Lee H, Arnaoutov A, Dasso M. Association of RanGAP to nuclear pore complex component, RanBP2/Nup358, is required for pupal development in Drosophila. Cell Rep 2021 37:110151.
- Aksenova V, Dasso M. Mapping paths through the nuclear pore complex. Nat Cell Biol 2022 24:6–7.
- Aksenova V, Arnaoutov A, Dasso M. Analysis of Nucleoporin Function Using Inducible Degron Techniques. Methods Mol Biol 2022 2502:129–150.
- Bley CJ, Nie S, Mobbs GW, Petrovic S, Gres AT, Liu X, Mukherjee S, Harvey S, Huber FM, Lin DH, Brown B, Tang AW, Rundlet EJ, Correia AR, Chen S, Regmi SG, Stevens TA, Jette CA, Dasso M, Patke A, Palazzo AF, Kossiakoff AA, Hoelz A. Architecture of the cytoplasmic face of the nuclear pore. Science 2022 376:6598.
- Gao S, Esparza M, Dehghan I, Aksenova V, Zhang K, Batten K, Ferretti MB, Begg BE, Cagatay T, Shay JW, García-Sastre A, Goldsmith EJ, Chen ZJ, Dasso M, Lynch KW, Cobb MH, Fontoura BMA. Nuclear speckle integrity and function require TAO2 kinase. Proc Natl Acad Sci U S A 2022 119:e2206046119.
Collaborators
- Beatriz Fontoura, PhD, University of Texas Southwestern Medical Center, Dallas, TX
- Amnon Harel, PhD, Azrieli Faculty of Medicine, Bar-Ilan University, Safed, Israel
- Lindsey Hayes, MD, Johns Hopkins University School of Medicine, Baltimore, MD
- André Hoelz, PhD, Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, CA
- Brian C. Oliver, PhD, Laboratory of Biochemistry and Genetics, NIDDK, Bethesda, MD
- Lionel Pintard, PhD, Institut Jacques Monod, Université Paris Diderot, CNRS, Paris, France
- Jeffrey Rothstein, MD, Brain Science Institute, Johns Hopkins University School of Medicine, Baltimore, MD
- Thomas U. Schwartz, PhD, Massachusetts Institute of Technology, Cambridge, MA
- Mihaela Serpe, PhD, Section on Cellular Communication, NICHD, Bethesda, MD
- Alexander Strunnikov, PhD, Guangzhou Institutes of Biomedicine and Health, Guangzhou, China
- Katharine Ullman, PhD, Huntsman Cancer Institute, University of Utah, Salt Lake City, UT
- Weidong Yang, PhD, Temple University, Philadelphia, PA
Contact
For more information, email mdasso@helix.nih.gov or visit https://sccr.nichd.nih.gov.