Mechanisms of Synapse Assembly and Homeostasis

- Mihaela Serpe, PhD, Head, Section on Cellular Communication
- Peter Nguyen, Biological Laboratory Technician
- Tae Hee Han, PhD, Staff Scientist
- Rosario Vicidomini, PhD, Research Fellow
- Wen Chieh Hsieh, PhD, Visiting Fellow
- Tho Huu Nguyen, PhD, Visiting Fellow
- Debabrata Sinha, PhD, Visiting Predoctoral/Postdoctoral Fellow
- SiJia Chen, BSc, Postbaccalaureate Fellow
- Thomas Brody, PhD, Special Volunteer
The purpose of our research is to understand the mechanisms of synapse development and homeostasis. The chemical synapse is the fundamental nervous-system communication unit, which connects neurons to one another and to non-neuronal cells, and is designed to mediate rapid and efficient transmission of signals across the synaptic cleft. This transmission forms the basis for the biological computations that underlie and enable our complex behavior. Crucial to the function is the ability of a synapse to change its properties, so that it can optimize its activity and adapt to the status of the cells engaged in communication and/or to the larger network comprising them. Consequently, synapse development is a highly orchestrated process coordinated by intercellular communication between the pre- and postsynaptic compartments and by neuronal activity itself. Our long-term goal is to elucidate the molecular mechanisms that regulate the formation of functional synapses during development and that fine-tune them during plasticity and homeostasis. We focus on four key processes in synaptogenesis: (1) trafficking of components to the proper site; (2) organizing those components to build synaptic structures; (3) maturation of the synapse to optimize its activity; and (4) homeostatic mechanisms that restore synapse activity to a set point after perturbations. We address the molecular mechanisms underlying these processes using a comprehensive set of approaches, which include genetics, biochemistry, molecular biology, super-resolution imaging, and electrophysiology recordings in live animals and in reconstituted systems.
Because of its many advantages, we study these events in a powerful genetics system, Drosophila melanogaster, and use the neuromuscular junction (NMJ) as a model for glutamatergic synapse development and function. The fact that individual NMJs can be reproducibly identified from insect to insect and are easily accessible for electrophysiological and optical analysis makes them uniquely suited for in vivo studies on synapse assembly, growth, and plasticity. In addition, the richness of genetic manipulations that can be performed in Drosophila permits independent control of individual synaptic components in distinct cellular compartments. Importantly, the fly NMJ relies entirely on kainate-type receptors, a family of ionotropic glutamate receptors that impact synaptic transmission and neuronal excitability in the mammalian central nervous system but remain poorly understood. The Drosophila NMJ can thus be used to analyze and model defects in the structural and physiological plasticity of glutamatergic synapses, which are associated with a variety of human pathologies, from learning and memory deficits to autism. Drosophila has long served as a source of insight into human genetics, development, and disease, and the basic discoveries of our laboratory in the fly are likely to serve our overarching goal of understanding how chemical synapses are assembled and sculpted during development and homeostasis.
Cellular diversity in the Drosophila third instar larval ventral cord revealed by single-cell transcriptomics
In flies as in vertebrates, neuronal activity induces input-specific changes in synaptic strength; at the larval NMJ, the postsynaptic sensitivity is primarily modulated via synapse-specific recruitment of postsynaptic glutamate receptors. Robust homeostatic mechanisms keep synapses within an appropriate dynamic range, such that the evoked potentials measured in the muscle remain constant from embryo to third instar larvae. Reduced postsynaptic sensitivities (i.e., reduced glutamate receptor activity) trigger a compensatory increase in quantal content (QC), i.e., the number of vesicles released by the motor neuron (MN), which is referred to as presynaptic homeostatic potentiation (PHP). To learn how MNs respond to various signals from muscles or compensate for perturbations in NMJ activity, we took a transcriptomic approach. We performed single-cell RNA sequencing (scRNA-Seq) of larval ventral nerve cords (VNCs), the fly equivalent of the mammalian spinal cord. However, MNs represent a relatively small fraction (less than 2%) of the cells within the larval VNC. To recognize the MN–specific transcriptomes and to begin comparing them under different conditions/genotypes, we first had to assemble a larval VNC atlas that captured and defined the cellular diversity within the Drosophila third instar larvae. During this process, we adapted and developed new protocols for dissociating single cells from fly larvae, then assembled a custom multistage analysis pipeline that integrates modules contained in different R packages, in order to ensure flexible, high-quality RNA-Seq data analysis. The work was conducted with support from the NICHD Genomics Core and in collaboration with Steve Coon, Fabio Rueda Faucz, and James Iben.
We dissected third instar larvae VNCs, dissociated the cells, and sequenced about 31,000 high-quality single cells. Using un-supervised clustering algorithms, we clustered the cells into distinct populations. We then assigned the populations to specific cell types using known markers. Through a series of reiterative processes, we first identified eight different glia subtypes, each with distinct metabolic pathways. Secondly, based on the expression of neuroblast genes and the temporal determinant genes, we revealed a developmental trajectory leading from neural precursors to newborn neurons. We also detected novel, differentially expressed genes along this trajectory. Thirdly, we identified over 40 types of clearly differentiated interneurons, each expressing unique combinations of transcription factors and neurotransmitters (VGlut, Gad1, VAChT, DAT). We found that most interneuron subtypes express many GPCRs (G protein–coupled receptors) and cell-recognition molecules, suggesting that many sensory modalities converge onto single cells to elicit specific motor functions/behaviors. We also identified a large MN cluster in which all cells express VGlut (encoding a glutamate transporter) but not the other neurotransmitter transporters or markers (Gad1, VAChT, DAT). As expected, the MN transcriptomes are enriched in futsch (encoding a microtubule-binding protein involved in the formation of synaptic boutons at the neuromuscular junctions), proctolin (proc, encoding a neurohormone that modulates NMJ function through unknown mechanisms), and target of wit (twit, a BMP transcriptional target previously implicated in NMJ function).
The fly larval/primary MNs are probably the most studied and best understood neurons, as their accessibility and stereotyped morphology has facilitated in-depth studies for almost 50 years. However, to date there is no systematic transcriptome analysis or class-specific characterization the larval MNs. Each abdominal hemisegment has 30 body-wall muscles innervated by about 36 larval MNs of four classes: type Ib (tonic) and type Is (phasic) glutamatergic MNs, type II octopaminergic and glutamatergic, and type III peptidergic neurons. To improve our ability to examine the heterogeneity within the MN cluster isolated in our scRNA-seq data, we marked the MNs with a twit-Gal4 promoter (and UAS-nls-GFP), dissociated VNCs from twit-GFP third instar larvae, and FACS–sorted the GFP–positive MNs. We accomplished the FACS sorting of the small Drosophila MNs in collaboration with Dragan Maric. The additional enrichment step allowed us to generate high quality scRNA-Seq data for over 1,200 MNs. This large pool of MNs was then subdivided in 28 clusters. Several of the clusters are very well isolated and correspond to neurosecretory cells (expressing orcokinin, leukokonin, or GPa), type II MNs (expressing Tdc2 and Vmat) and type III/peptidergic neurons (expressing the neurohormone bursicon and the neuropeptide crustacean cardioactive peptide, CCAP). Our dataset reveals new markers for these types of neurons. For example, the AMPA–type glutamate receptor subunits GluRIA and GluRIB are primarily expressed in type II and type III neurons, suggesting that the modulatory activity of these neurons is regulated by glutamatergic input.
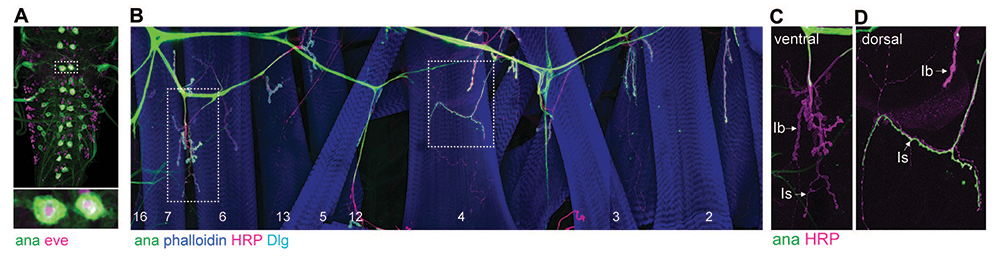
Drosophila has two type I MNs: tonic I-b neurons (with large synaptic boutons), which innervate single, dedicated muscles; and phasic I-s neurons, with small boutons and innervation spanning up to 7–8 muscles. Each larval hemisegment contains two I-s MNs, one projecting dorsally and one ventrally, and about 30 type I-b MNs, organized in distinct bundles that innervate subsets of body-wall muscles. Among the 28 MN clusters, two adjacent ones correspond to I-s MNs; they both express DIP-alpha (encoding a cell-surface molecule of the immunoglobulin superfamily), with the transcription factor gene eve marking the dorsally projecting I-s cluster. We found that anachronism (ana, encoding a secreted glycoprotein) specifically marks dorsally projecting I-s and confirmed this restricted expression using an ana-Gal4 (CRIMIC) line (Figure 1). We also recognized different I-b MN bundles, projecting dorsally, ventrally, or laterally. We already have a distinct set of markers so that all types of larval MNs can be unequivocally identified, irrespective of genetic background or synapse activity. Current work focuses on increasing the granularity of MN transcriptomic analysis and describing each of the MN bundles that innervate the larval body-wall muscles.
Click image to view.
Figure 1. ana, encoding a secreted glycoprotein, is specifically expressed in dorsal type Is motor neurons.
Confocal images of ventral nerve cord (A) or muscle fields (B-D) from third instar larvae illustrating specific expression of ana (ana-Gal4>UAS-CD4::GFP)(green) in only two type-Is motor neurons per larval segment. The ventral cord (A) is co-labeled for eve (magenta), a transcription factor that marks dorsally projecting motor neurons. The NMJ fields are stained for phalloidin (blue) (B), which marks the body-wall muscles, for Horseradish Peroxidase (HRP) (magenta), which labels an epitope of the neuron cell membrane and for Discs-large (Dlg) (cyan), a scaffold protein that accumulates predominantly around the type Ib terminals. The membrane-attached GFP, driven by the ana-Gal4 promoter, specifically labels dorsal (D) but not ventral (C) type Is motor neurons.
Our studies on the transcriptomes of larval MNs together with the assembly of a larval VNC atlas have already uncovered new molecules critical for synapse development and function. One example of a novel glutamate receptor will be discussed below. In addition, the larval VNC will provide a valuable resource for future studies on neuronal development and behavior.
A novel kainate receptor subunit modulates basal neurotransmission and homeostasis at the Drosophila NMJ
In flies, as in humans, the interplay between different postsynaptic receptor subtypes with different channel properties controls synapse strength and plasticity. The Drosophila NMJ utilizes two types of postsynaptic receptors, types A and B, which contain either GluRIIA or GluRIIB subunits, plus GluRIIC, GluRIID and GluRIIE; these receptors mediate the postsynaptic response to neurotransmitter. In addition, a presynaptic autoreceptor containing the KaiR1D subunit controls basal neurotransmission. At excitatory synapses, autoreceptors provide a feedback mechanism that modulates neurotransmitter release and ensures stable neuronal network activities. Phylogenetic analysis indicates that all these pre- and postsynaptic glutamate receptor subunits are closely related to the vertebrate kainate receptors, although the Drosophila receptors have strikingly different ligand-binding profiles. Like vertebrate kainate receptors, Drosophila kainate-type receptors are modulated by a member of the Neto (Neuropilin and Tolloid-like) family of auxiliary proteins (see below).
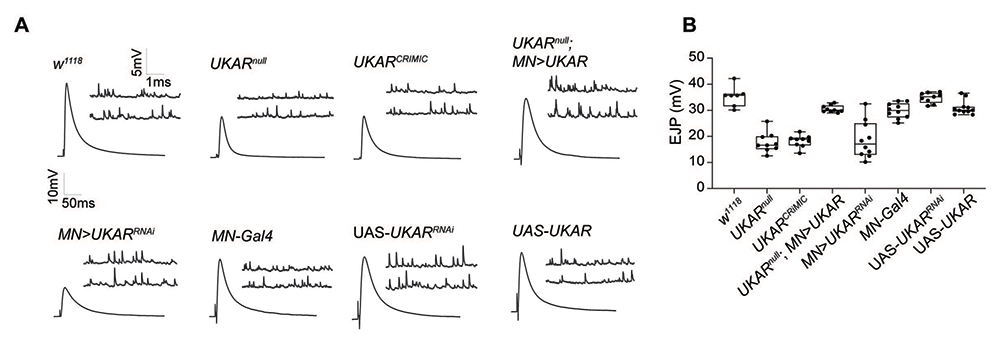
While characterizing larval MN transcriptomes, we noted that all type I MNs show high expression of a novel gene, CG11155, predicted to encode a kainate-type receptor subunit. The transcript was also abundant in larval interneurons (our dataset) as well as in many cells of the adult fly brain (recently published); this unusual, ubiquitous expression prompted us to refer to this gene as ubiquitous kainate receptor (ukar). To search for the role for UKAR during MNs development and function, we used CRISPR/Cas9 and RNAi methodologies to generate null mutants and tissue-specific knock downs. We found that UKAR functions in MNs to ensure normal basal neurotransmission; in the absence of UKAR, basal neurotransmission is reduced to half of the control (Figure 2), a loss-of-function phenotype that is reminiscent of the KaiRIDnull mutants. Given that glutamate receptors function as heterotetramers, i.e., usually dimers of dimers, our data suggest that KaiRID and UKAR represent the two subunits of a presynaptic autoreceptor that controls basal neurotransmission at the larval NMJ.
Figure 2. UKAR, a novel kainate receptor subunit, modulates basal neurotransmission at larval NMJ.
A. Representative traces for miniature excitatory junction potentials (mEJPs) (which represent the postsynaptic response to the spontaneous release of single synaptic vesicles) and evoked excitatory junction potentials (EJPs) from muscle 6 of the indicated genotypes.
B. Bar graph illustrating the EJP amplitudes in various genotypes.
Reduction in postsynaptic receptor activities causes reduced frequency and amplitude of miniature excitatory junction potentials (mEJPs), but such NMJs have normal evoked potentials (EJPs) owing to a compensatory increase in neurotransmitter release, a phenomenon referred to as presynaptic homeostatic potentiation (PHP). For example, application of sub-blocking concentrations of philanthotoxin (PhTx), a polyamine toxin derived from wasp venom, to semi-intact larval preparations triggers a fast reduction in quantal size and an increase in quantal content (QC), so that the basal neurotransmission recovers within minutes. Loss of presynaptic KaIRID (or Neto) renders such NMJs unable to express PHP. Likewise, loss of UKAR (in ukarnull mutants) or knockdown of ukar specifically in MNs, induced loss of ability to express PHP at these NMJs: upon toxin application the mini amplitudes were reduced but the basal neurotransmission never recovered. Expression of a ukar transgene in larval MNs effectively restored basal neurotransmission and PHP to normal levels. Together, our results indicate that KaiRID and UKAR share multiple activities at larval NMJ and that they probably function as a heterodimer autoreceptor.
Neto, a highly conserved auxiliary subunit that modulates kainate-type receptors
We previously discovered that an obligatory auxiliary protein, Neto, is absolutely required for clustering of postsynaptic glutamate receptors and for NMJ functionality. Neto belongs to a family of highly conserved auxiliary proteins that share an ancestral role in the formation and modulation of glutamatergic synapses. Vertebrate Neto1 and Neto2 were shown to modulate the properties of selective glutamate receptors, in particular the kainate-type receptors. Our previous investigations unveiled essential roles for Drosophila Neto during NMJ development and strongly support the notion that trafficking of both postsynaptic and the presynaptic glutamate receptors, their synaptic recruitment and stabilization, and their function are tightly regulated by Neto. We found that the fly Neto directly engages the glutamate receptor complexes, as well as other intracellular and extracellular proteins, to selectively regulate the distribution of postsynaptic glutamate receptor subtypes, the recruitment of postsynaptic proteins, and the organization of postsynaptic structures. In recent studies, we focused on how Neto modulates the gating properties of post- and pre-synaptic receptors.
Drosophila neto encodes two isoforms, Neto-α and Neto-β, which share the extracellular and transmembrane domains but have distinct intracellular parts. Neto-β is the predominant isoform at the NMJ and functions in the muscle to recruit glutamate receptors and other postsynaptic components. Neto-α acts predominantly in the motor neurons to ensure normal basal neurotransmission. To study the biophysical properties of Drosophila NMJ receptors, we are using fast agonist application on outside-out patches from HEK293 cells transfected with various combinations of receptors with or without Neto isoforms/variants. Neto is critical for the functional reconstitution of both postsynaptic receptors (type-A: GluRIIA/C/D/E and type-B: GluRIIB/C/D/E): 100% of the more than 100 patches yielded no currents in the absence of Neto proteins. Furthermore, the two isoforms, Neto-α and Neto-β, differentially modulate the desensitization and deactivation rates for postsynaptic receptors/channels. The desensitization rates are similar for type-A or type-B channels in complexes with Neto-β or Neto-ΔCTD (which lacks any intracellular domain), but are significantly reduced when Neto-α is co-transfected with either postsynaptic receptor complex. Neto is absolutely required for the function of type-A and type-B receptors, but not for the KaiR1D homotetramer channels. When transfected by itself, KaiRID forms rapidly activating and desensitizing channels. When KaiR1D was co-transfected with Neto proteins, the receptor's expression increased (from 36% to 61.8% success rate for KaiR1D/Neto-α complexes) and the gating kinetics were altered. We found non-significant changes in the deactivation rates, but the desensitization time constants increased two-fold from KaiRID alone to KaiRID in combination with Neto-α, Neto-β, or Neto-ΔCTD. The CUB1 domain of Neto proteins is absolutely required for any Neto-dependent modulation. This is probably because this domain was predicted by Cryo-EM studies to form strong interactions, an anchor point, with the amino-terminal domain of the receptor complexes. The intracellular domains of Neto had differential effects on various receptors/channels. Pre-treatment with Concanavalin A increases the single-channel open times for all postsynaptic and presynaptic receptors, whereas extracellular philanthotoxin blocks the channels to various extents. These channels are differentially regulated by addition of intracellular polyamines (spermine) at physiological concentration, which changes the rectification profiles for each channel. Our studies reveal that Neto is not only required for the function of selected channels but also increases the diversity of the receptor properties.
Novel split methodology to image, track, and reconstitute complex proteins
Synaptic proteins such as Neto and glutamate receptors are notoriously difficult to track and study because of their low abundance and the high density of functional domains. Studies using transgenic lines that express selective tagged isoforms/variants often cannot generate definitive conclusions owing to heterologous promoters and overexpression artifacts. To facilitate structure-function studies and accomplish reliable detection of low-abundant synaptic proteins in different tissues, we turned to a recently described cell-biology tool, the ALFA system. The system consists of a synthetically designed epitope tag of only 14 amino acids, the ALFA tag (AT), with no homology in the animal kingdom, and a nanobody (NbALFA) that binds to ALFA–tagged proteins with picomolar affinities. The high affinity of ALFA tag/NbALFA binding and the intrabodies capabilities of NbALFA (that is binding ALFA–tagged proteins when expressed in living cells) have prompted the development of a variety of in vitro cell-biology applications, from super-resolution to live detection of tagged proteins.
To probe whether the methodology is suitable for in vivo application, we chose a case-study protein, Drosophila Neurexin-1 (Nrx-1). Neurexins are key adhesion proteins that coordinate extracellular and intracellular synaptic assembles. Neurexins are also notoriously difficult to track and study because of their low abundance and the high density of functional domains. These proteins are crucial for synapse assembly and function; however, the role of some of their domains (for example the C-terminal PDZ binding motifs) has only been inferred from in vitro studies. Guided by phylogenetic analysis and secondary structure prediction, we generated ALFA–tagged Nrx-1 variants, including an endogenously tagged Nrx-1-AT allele, which is indistinguishable from the wild-type control, and a NrxDPDZ-AT allele that resembles the Nrx-1null mutant. Using a combination of classic genetics and cell biology and electrophysiology approaches, we found that Nrx-1-AT NMJs have normal morphology and function, whereas the NrxDPDZ-AT mutants have smaller NMJs with much reduced basal neurotransmission, reminiscent of Nrx-1null mutant. Similar to untagged Nrx-1, endogenously edited Nrx-1-AT localizes at presynaptic sites. Remarkably, the ALFA system enabled detection of endogenous Nrx-1-AT in only one immunohistochemistry step, using the monovalent binder NbALFA conjugated to two fluorophores (FluoTag-X2 anti-ALFA). Using cytosolic NbALFA-mScarlet intrabody, we also tracked live Nrx-1 transport along the motor neuron axons and described fast anterograde and slow retrograde synaptic transport vesicles. These data confirm the expectation that the ALFA system ensures high-affinity binding, linear (monovalent) signals with respect to target molecule, with no amplification by polyclonal secondaries, and virtually no background in animal tissues.
In addition, we found that the PDZ binding motif is key to Nrx-1 in vivo surface expression and synaptic localization: the NrxDPDZ-AT variant was trapped in the ER, unable to traffic to the cell surface. This explained why the NrxDPDZ-AT allele had NMJ defects similar with the Nrx-1null mutant. Given that the ALFA system is very compact and has high binding affinity, both inside and outside the cells, we next investigated whether the system could deliver the missing PDZ binding motif in trans, facilitating the reconstitution of functional Nrx-1. To this end, we generated a genetically encoded NbALFA–PDZ binding motif chimera (UAS-Nb-PDZ) and expressed it in the NrxDPDZ-AT neurons. The resulting animals were viable and fertile and had normal NMJ morphology and function, indicating that a PDZ–binding motif provided in trans fully restored the synaptic localization and function of NrxDPDZ-AT. The ability to use the ALFA system as a split system to reconstitute and track functional proteins in vivo opens up a new realm of possibilities for functional studies in specific cells/tissues and during defined developmental windows. We anticipate that this methodology will pave the way towards dissecting functional domains of complex proteins in vivo.
Publications
- Vicidomini R, Serpe M. Local BMP signaling: a sensor for synaptic activity that balances synapse growth and function. Curr Topics Dev Biol 2022 50:211–246.
- Nguyen TH, Vicidomini R, Choudhury S, Coon SL, Iben J, Brody T, Serpe M,. Single-cell RNA sequencing analysis of the Drosophila larval ventral cord. Curr Protoc 2021 e38.
- Vicidomini R, Nguyen TH, Choudhury S, Brody T, Serpe M. Assembly and exploration of a single cell atlas of the Drosophila larval ventral cord. Identification of rare cell types. Curr Protoc 2021 e37.
- Han TH, Vicidomini R, Ramos CI, Wang Q, Nguyen P, Jarnik M, Lee CH, Stawarski M, Hernandez RX, Macleod GT, Serpe M. Neto-alpha controls synapse organization and homeostasis at the Drosophila neuromuscular junction. Cell Rep 2020 32:107866.
- Nguyen TH, Han TH, Newfeld SJ, Serpe M. Selective disruption of synaptic BMP signaling by a Smad mutation adjacent to the highly conserved H2 helix. Genetics 2020 216:159–175.
Collaborators
- Steven Coon, PhD, Molecular Genetics Core, NICHD, Bethesda, MD
- James Iben, PhD, Molecular Genetics Core, NICHD, Bethesda, MD
- Gregory T. Macleod, PhD, Florida Atlantic University, Jupiter, FL
- Dragan Maric, PhD, Flow and Imaging Cytometry Core, NINDS, Bethesda, MD
- Stuart Newfeld, PhD, School of Life Sciences, Arizona State University, Tempe, AZ
- Felipe Opazo, PhD, Center for Biostructural Imaging of Neurodegeneration, Göttingen, Germany
- Fabio Rueda-Faucz, PhD, Molecular Genetics Core, NICHD, Bethesda, MD
Contact
For more information, email mihaela.serpe@nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/Serpe.