Chemosensory Coding and Decoding by Neuron Ensembles

- Mark Stopfer, PhD, Head, Section on Sensory Coding and Neural Ensembles
- Zane Aldworth, PhD, Staff Scientist
- Alejandra Boronat Garcia, PhD, Postdoctoral Fellow
- Yu-Shan Hung, PhD, Postdoctoral Fellow
- Subhasis Ray, PhD, Postdoctoral Fellow
- Bo-Mi Song, PhD, Postdoctoral Fellow
- Kui Sun, MD, Technician
- Brian Kim, BS, Graduate Student
- Leah Pappalardo, BS, Graduate Student
All animals need to know what is going on in the world around them. Brain mechanisms have thus evolved to gather and organize sensory information to build transient and sometimes enduring internal representations of the environment.
Using relatively simple animals and focusing primarily on olfaction and gustation, we combine electrophysiological, anatomical, behavioral, computational, optogenetic, and other techniques to examine the ways in which intact neural circuits, driven by sensory stimuli, process information. Our work reveals basic mechanisms by which sensory information is transformed, stabilized, and compared, as it makes its way through the nervous system.
We use three species of insects, each with specific and interlocking experimental advantages, as our experimental preparations: locusts, moths, and fruit flies. Compared with the vertebrate, the insect nervous system contains relatively few neurons, most of which are readily accessible for electrophysiological study. Essentially intact insect preparations perform robustly following surgical manipulations, and insects can be trained to provide behavioral answers to questions about their perceptions and memories. Ongoing advances in genetics permit us to target specific neurons for optogenetic or electrophysiological recording or manipulations of activity. Furthermore, the relatively small neural networks of insects are ideal for tightly constrained computational models that test and explicate fundamental circuit properties.
Response heterogeneity and adaptation in olfactory receptor neurons
The olfactory system, consisting of relatively few layers of neurons, with structures and mechanisms that appear repeatedly in widely divergent species, provides unique advantages for the analysis of information processing by neurons. Olfaction begins when odorants bind to olfactory receptor neurons, triggering them to fire patterns of action potentials. Recently, using new electrophysiological recording tools, we found that the spiking responses of olfactory receptor neurons are surprisingly diverse and include powerful and variable history dependencies. Single, lengthy odor pulses elicit patterns of excitation and inhibition that cluster into four basic types. Different response types undergo different forms of adaptation during lengthy or repeated stimuli. A computational analysis showed that such diversity of odor-elicited spiking patterns helps the olfactory system efficiently encode odor identity, concentration, novelty, and timing, particularly in realistic environments.
Feedback inhibition and its control in an insect olfactory circuit
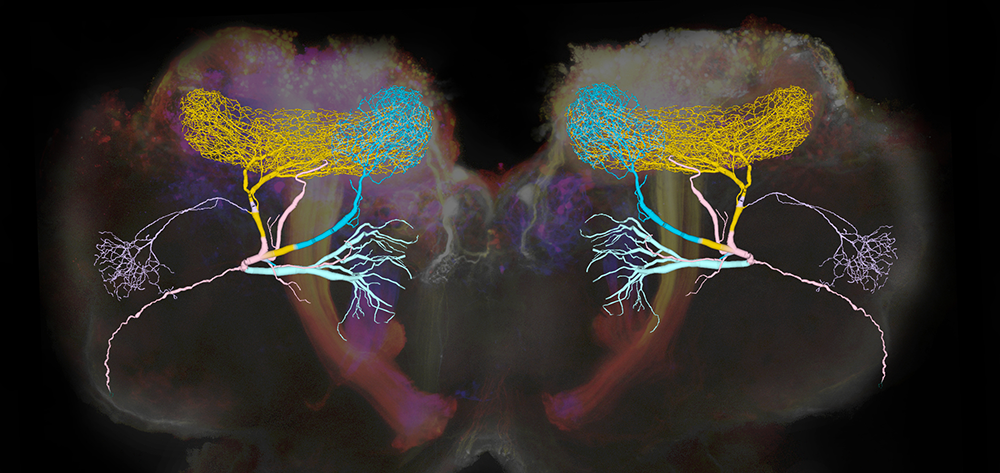
Inhibitory neurons play critical roles in regulating and shaping olfactory responses in vertebrates and invertebrates. In insects, such roles are performed by relatively few neurons, which can be interrogated efficiently, revealing fundamental principles of olfactory coding. With electrophysiological recordings from the locust and a large-scale biophysical model, we analyzed the properties and functions of the giant GABAergic neuron (GGN), a unique neuron that plays a central role in structuring olfactory codes in the locust brain (Figure 1). Analysis of our in vivo recordings and simulations of our model of the olfactory network suggest that the GGN extends the dynamic range of Kenyon cells (high-order neurons in a brain area analogous to the vertebrate piriform cortex, which fire spikes when the animal is presented with an odor pulse), which leads us to predict the existence of a yet undiscovered olfactory pathway. Our analysis of GGN–intrinsic properties, inputs, and outputs, in vivo and in silico, reveals basic new features of this critical neuron and the olfactory network that surrounds it. Together, results of our in vivo recordings and large-scale realistic computational modeling provide a more complete understanding of how different parts of the olfactory system interact.
Figure 1. Giant GABAergic neurons regulate olfactory responses in the locust brain.
The composite image shows the structure of a compartmental computational model of the giant GABAergic neurons (GGNs) superimposed on dextran-dyed mushroom bodies in the locust brain. Different branches of GGN are shown in different colors. GGNs, only one on each side of the brain, regulate the firing of tens of thousands of olfactory neurons through feedback inhibition.
Spatiotemporal coding of individual chemicals by the gustatory system
Four of the five major sensory systems (vision, olfaction, somatosensation, and audition) are thought to be encoded by spatiotemporal patterns of neural activity. The exception is gustation. Gustatory coding by the nervous system is thought to be relatively simple, i.e., every chemical (‘tastant’) is associated with one of a small number of basic tastes, and the presence of a basic taste, rather than the specific tastant, is represented by the brain. In mammals as well as insects, five basic tastes are usually recognized: sweet, salty, sour, bitter, and umami. The neural mechanism for representing basic tastes is unclear. The most widely accepted postulate is that, in both mammals and insects, gustatory information is carried through labeled lines of cells sensitive to a single basic taste, that is, in separate channels from the periphery to sites deep in the brain. An alternative proposal is that the basic tastes are represented by populations of cells, with each cell sensitive to several basic tastes.
Testing these ideas requires determining, point-to-point, how tastes are initially represented within the population of receptor cells and how this representation is transformed as it moves to higher-order neurons. However, it has been highly challenging to deliver precisely timed tastants while recording cellular activity from directly connected cells at successive layers of the gustatory system. Using a new moth preparation, we designed a stimulus and recording system that allowed us to fully characterize the timing of tastant delivery and the dynamics of the tastant-elicited responses of gustatory receptor neurons and their monosynaptically connected second-order gustatory neurons, before, during, and after tastant delivery.
Surprisingly, we found no evidence consistent with a basic taste model of gustation. Instead, we found that the moth’s gustatory system represents individual tastant chemicals as spatiotemporal patterns of activity distributed across the population of gustatory receptor neurons. We further found that the representations are transformed substantially, given that many types of gustatory receptor neurons converge broadly upon follower neurons. The results of our physiological and behavioral experiments suggest that the gustatory system encodes information not about basic taste categories but rather about the identities of individual tastants. Furthermore, the information is carried not by labeled lines but rather by distributed, spatiotemporal activity, which is a fast and accurate code. The results provide a dramatically new view of taste processing.
Argos: a toolkit for tracking multiple animals in complex visual environments
Understanding how neural mechanisms drive behaviors often requires a rigorous analysis of those behaviors, a task that can be arduous and time-consuming. Many recent software utilities can automatically track animals in homogeneous, uniformly illuminated scenes with constant backgrounds, using traditional image-processing algorithms, and recently, neural networks have improved animal tracking and pose-estimation software. However, these tools are optimized to analyze experiments in unrealistically simplified laboratory environments, for example, by using thresholding steps to detect contiguous binary objects in a video frame, and then using this information to automatically train a neural network to identify the animals. The thresholding-based detection can fail if the background changes over time or is cluttered with objects of size and contrast similar to target animals. Some multi-animal pose-estimation tools can identify individual animals even with inhomogeneous backgrounds, but such tools require either large manually annotated datasets for training or are practically limited to a few animals. We focused on the task of tracking multiple animals, especially in inhomogeneous or changing environments, without visual identification or pose estimation.
To facilitate capturing video, training neural networks, tracking several animals, and reviewing tracks, we developed Argos, a software toolset that incorporates both classical and neural net–based methods for image processing, and provides simple graphical interfaces to control parameters used by the underlying algorithms. Argos includes tools for compressing videos based on animal movement, for generating training sets for a convolutional neural network (CNN) to detect animals, for tracking many animals in a video, and for facilitating review and correction of the tracks manually, with simple graphical user interfaces. Argos can help reduce the amount of video data to be stored and analyzed, speed up analysis, and permit analyzing difficult and ambiguous conditions in a scene. Thus, Argos supports several approaches to animal tracking suited for varying recording conditions and available computational resources. Together, these tools allow the recording and tracking of movements of many markerless animals in inhomogeneous environments over many hours. The tools thus provide many benefits to researchers.
Identification and analysis of odorant receptors expressed in two main olfactory organs: antennae and palps of a model organism, the locust Schistocerca americana
Olfaction allows animals to detect, identify, and discriminate among hundreds of thousands of odor molecules present in the environment. It requires a complex process to generate the high dimensional neural representations needed to characterize odorant molecules, which have different sizes, shapes and electrical charges, and are often organized into chaotic and turbulent odor plumes. Understanding the anatomical organization of the olfactory system at the cellular and molecular levels has provided important insights into the coding mechanisms underlying olfaction, and studies performed in insects have contributed substantially to our knowledge of odor processing. Further, mechanisms that allow the olfactory system to generate representations for odors have been shown to be widely conserved among very divergent species. Extending our understanding of olfaction requires knowledge of the molecular and structural organization of the olfactory system.
Odor sensing begins with olfactory receptor neurons (ORNs), which express odorant receptors (ORs). In insects, ORNs are housed, in varying numbers, in olfactory sensilla. Because the organization of ORs within sensilla affects their function, it is essential to identify the ORs they contain. Using RNA sequencing, we identified 179 putative ORs in the transcriptomes of the two main olfactory organs, antenna and palp, of the locust Schistocerca americana. Quantitative expression analysis showed most putative ORs (140) are expressed in antennae, while only 31 are in the palps. Further, one OR was detected only in palps, and seven are expressed differentially by sex. An in situ analysis of OR expression revealed at least six classes of sensilla in the antenna. A phylogenetic comparison of OR–predicted protein sequences revealed homologous relationships among two other Acrididae species. Our results provide a foundation for understanding the organization of the first stage of the olfactory system in S. americana, a well studied model for olfactory processing.
Additional Funding
- NICHD Career Development Award to Dr. Bo-Mi Song
Publications
- Ray S, Aldworth Z, Stopfer M. Feedback inhibition and its control in an insect olfactory circuit. eLife 2020 9:e53281.
- Gupta N, Singh SS, Stopfer M. Oscillatory integration windows in neurons. Nat Commun 2016 7:13808.
- Reiter S, Campillo Rodriguez C, Sun K, Stopfer M. Spatiotemporal coding of individual chemicals by the gustatory system. J Neurosci 2015 35:12309–12321.
- Ray S, Stopfer M. Argos: a toolkit for tracking multiple animals in complex visual environments. Methods Ecol Evolution 2022 13:3:585–595.
- Aldworth ZN, Stopfer M. Insect neuroscience: filling the knowledge gap on gap junctions. Curr Biol 2022 32(9):R420–R423.
Collaborators
- Maxim Bazhenov, PhD, Howard Hughes Medical Institute, The Salk Institute for Biological Studies, La Jolla, CA
Contact
For more information, email stopferm@mail.nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/stopfer or https://dir.ninds.nih.gov/Faculty/Profile/mark-stopfer.html.