The Biological Impact of Transposable Elements

- Henry L. Levin, PhD, Head, Section on Eukaryotic Transposable Elements
- Angela Atwood-Moore, BA, Senior Research Assistant
- Abhishek Anand, PhD, Postdoctoral Fellow
- Paul Atkins, PhD, Postdoctoral Fellow
- Hyo Won Ahn, PhD, Visiting Fellow
- Feng Li, PhD, Visiting Fellow
- Rakesh Pathak, PhD, Visiting Fellow
- Abigail Burkhart, BS, Postbaccalaureate Fellow
- Kyla Roland, BA, Postbaccalaureate Fellow
Long Terminal Repeat (LTR) retrotransposons are highly abundant and have evolved into ubiquitous families of elements that multiply through cycles of particle formation, reverse transcription, transport to the nucleus, and integration. Some families of LTR–retrotransposons acquired envelope proteins, an addition that transformed the elements into infectious retroviruses. This close relationship makes LTR retrotransposons ideal models for studying the molecular mechanisms responsible for retrovirus replication. The transposable elements (TEs) of model organisms, such as yeast, are particularly well suited to address the dynamics and global impact of their replication. We study LTR retrotransposons of the fission yeast (Schizosaccharomyces pombe) to determine how integration sites are selected and to understand how patterns of integration impact the physiology of the cell. In past work, we found that integration of LTR retrotransposons in S. pombe alters gene expression and adapts cells to environmental stress. It is through adaptation that we believe TEs form gene-regulatory networks. We also study how HIV-1 integration sites are selected.
In humans, TEs represent 50% of genomic sequences. The dominant families of TEs are Long INterspersed Element-1 (LINE-1 or L1), which constitutes 17% of the genome, and Alu Short Interspersed Elements (SINEs), which are mobilized by L1 and constitute 10% of the genome. Given that TEs make up half of the human genome, it is not surprising that their regulatory features are abundant sources of tissue-specific promoter activity and are critical building blocks of gene-regulatory networks. Although the vast majority of TEs have lost mobility, each genome retains approximately 100 active copies. As a result, genome studies of human populations reveal many thousands of polymorphic TEs. Our goal is to determine the role of these genetic variants in health and disease.
Identification of an integrase-independent pathway of retrotransposition
Despite the central role of integration in the propagation of retroviruses, important questions remain about residual insertions that occur in the absence of integrase (IN) activity. Mutations in the catalytic residues of HIV-1 IN produce residual infectious titers, typically with a 3 to 4-log reduction. However, in continuous cultures of HIV-1 lacking IN activity, insertion efficiency can be as high as 0.2–0.8% of a wild-type (WT) virus. These findings indicate that retroviruses possess a secondary, IN–independent pathway, which incorporates viral DNA into the host genome. Given that IN–independent infections could compromise the treatment of HIV-1 patients with IN inhibitors, it is important to identify the nature of this pathway.
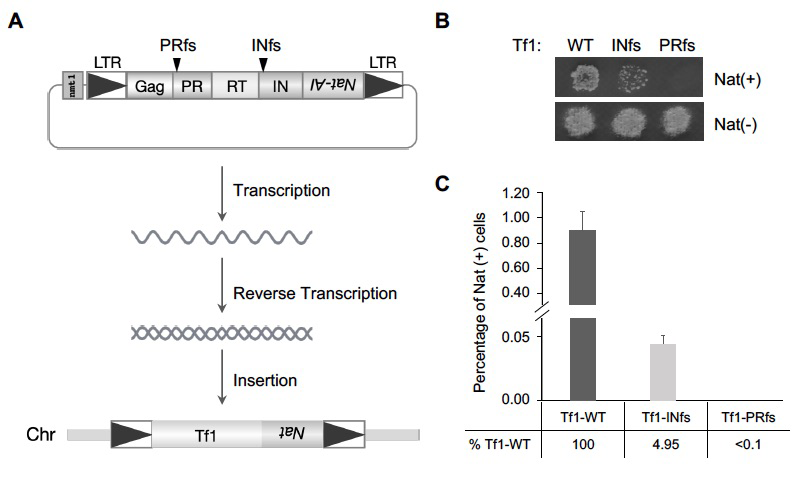
LTR retrotransposons are important models of retroviruses because of their structural and mechanistic similarities. Tf1 and Tf2 are extensively characterized LTR retrotransposons with high integration activity in S. pombe. Studies of Tf1 expressed with genetic markers demonstrate that the Gag protein, protease (PR), reverse transcriptase (RT), and IN all contribute to transposition. Importantly, the resulting integration is directed to specific RNA pol II promoters by the DNA–binding factor Sap1. To identify a model system that can be used to study the mechanisms of IN–independent insertion, we measured the insertion of Tf1 lacking IN activity. We performed an insertion assay with Tf1 encoding a frameshift mutation at the start of IN (Tf1-INfs) that blocks expression of IN without altering RT expression or cDNA synthesis. We found Tf1-INfs retained 4.95% of the insertion activity of Tf1-WT [Reference 1]. These results indicate that, in the absence of IN activity, Tf1 cDNA inserted into the host genome with surprising efficiency. Genome-wide insertion profiles of Tf1 lacking IN (Tf1-INfs) were significantly different from those of Tf1 expressing active IN. DNA logo analysis showed that the sequences downstream of the Tf1-INfs insertion sites had a prominent bias for ATAAC, and upstream flanks showed a preference of CAA. Interestingly, the downstream logo matches that of the primer binding site (PBS), an 11 bp sequence retained after reverse transcription on the 3′ end of the plus-strand cDNA. The CAA matches the last three base pairs of the poly purine tract (PPT), which is retained on the 3′ end of the minus-strand cDNA. The PBS and PPT preferences indicated that these single-stranded sequences contributed to insertion through homologous recombination (HR). If IN–independent insertions are directed to sites with homology to the PBS and PPT, we would expect that large numbers of insertions would occur at the 13 pre-existing copies of Tf2 that have PBS and PPT sequences identical to those of Tf1. By analyzing the raw downstream sequences, we found that approximately 70% of the IN–independent insertions occurred at homologous sequences within the pre-existing 5′ LTRs of Tf2s. Whole genome sequencing of these events revealed that the most common outcome of these insertions resulted in tandem copies of Tf1 and Tf2 elements.
Our data suggest that IN–independent insertion of Tf1 is likely mediated by a form of homologous recombination. To determine whether homologous recombination factors contribute to IN–independent insertion, we measured insertion frequencies of strains lacking mre11, rad50, nbs1, rad51, or rad52. The results revealed that the insertions occurred through Rad52–dependent single-strand annealing (SSA), as Rad51 was dispensable. The rad52–R45A mutation, which specifically abolishes the SSA activity of Rad52, significantly reduced the frequency of Tf1-INfs insertions and resulted in dissociation of Rad52 from Tf1 cDNA. These data indicate that Rad52 plays a critical role in IN–independent insertions by binding to the ends of the cDNA, causing recombination with sequences similar to PBS and PPT.
The efficiency of HR–mediated IN–independent insertion of Tf1 raised questions about whether this pathway has a biological function. Our efforts to determine whether IN–independent events occur naturally showed that cultures with continuing expression of WT Tf1 produced insertions that were predominantly IN–independent [Reference 1]. These data demonstrate that Tf1 possesses two efficient insertion pathways, one relying on IN and the other being IN–independent but requiring Rad52. Significantly, we found in previously published data of HIV-1 IN–independent insertions that five of 69 sites had strong similarity to the HIV-1 PBS. Together, these results indicate that homology-dependent SSA provides a significant pathway of IN–independent insertion.
Figure 1. Tf1 insertion takes place in the absence of integrase.
A. The diagram shows the strategy of monitoring Tf1 retrotransposition. A drug-resistant gene, nat, with artificial intron (nat-AI) is introduced into Tf1, and the integration of Tf1 into host chromosomes allows cells to grow on plates containing Nat. The black arrows indicate the frame shift (fs) sites of PR and IN respectively. LTR: long terminal repeat; PR: protease; RT: reverse transcriptase; IN: integrase; WT: wild-type.
B. Growth phenotypes of Tf1-WT, Tf1-INfs, and Tf1-PRfs on medium containing Nat after inducing Tf1 expression.
C. Quantitative transposition analysis Tf1-WT, Tf1-INfs, and Tf1-PRfs.
Retrotransposon insertions associated with risk of neurologic and psychiatric diseases
Mental disorders affected about 970 million people worldwide in 2017. In 2020, 21% of adults in the United States suffered from some form of mental illness. Consequently, these diseases cause great social and economic burden. Studies of identical twins show that the heritability of diseases such as attention-deficit hyperactivity disorder (ADHD), autism spectrum disorder (ASD), bipolar disorder (BIP), and schizophrenia is extremely high, ranging from 74% to 81%. Because of the complexity of the mammalian nervous system, the genetic and cellular etiology of such diseases remains largely unclear. Progress in genetic methodology has provided the potential to identify mechanisms that underlie the diseases. One approach that has successfully identified important disease loci is genome-wide association studies (GWAS). However, in the cases of neurologic and major psychiatric disorders, GWAS have identified large numbers of loci, each associated with small increases in risk. Importantly, there is extensive overlap of the loci that contribute to major psychiatric disorders, indicating that related molecular mechanisms may underlie distinct clinical phenotypes.
TASs (trait-associated single-nucleotide polymorphisms [SNPs]) of GWAS are genetic tags identifying a genomic region that contains the causal mutation(s), which lead to increased disease risk. Limits on the design of GWAS typically prevent such studies from identifying causal gene alleles. Thus, determining causal variants remains the most challenging and rate-limiting, but also the most important, step in defining the genetic architecture of diseases. The vast majority of GWAS TASs lie in intergenic or intronic regions and therefore do not alter coding sequence. For such SNPs to be causal they would likely have regulatory effects on transcription. Structural variants such as rearrangements, copy number variants, and transposable element (TE) insertions constitute a substantial and disproportionately large fraction of the genetic variants found to alter gene expression.
In humans, the dominant families of TEs are long interspersed element-1 (LINE-1 or L1) and Alu elements, which are short interspersed elements (SINEs) and are mobilized by L1. TEs readily alter gene expression because they have evolved various sequences that act on enhancers. Given that TEs make up approximately 45% of the human genome, it is not surprising that their regulatory features are abundant sources of tissue-specific promoter activity.
Relatively recent TE insertions can proliferate in the population and become common alleles. The 1000 Genomes Project described genetic variation of diverse human populations by sequencing whole genomes of 2,504 individuals. The extensive survey of genetic variation detected 17,000 polymorphic insertions of TEs, which have the potential to alter gene expression and affect common disease risk. There may be functional consequences of common TE insertion variants that affect common disease risk. Some common polymorphic TEs have been implicated at disease loci detected by GWAS. Common polymorphic Alu (short transposable elements) insertions occur disproportionately near disease loci of GWAS, underscoring the fact that Alu insertions are potential causative variants.
Given the difficulty in identifying genetic variants responsible for neurologic and psychiatric disorders and the regulatory capacity of TEs, we tested whether polymorphic TEs are potential causative variants of such diseases [Reference 2]. We analyzed 593 GWAS of neurologic and psychiatric diseases, which in total reported 753 TASs. From the 17,000 polymorphic TEs, we found that 76 were in linkage disequilibrium (LD) with TASs, indicating that the TEs were among the variants with the potential to be causative. We extended our analysis by evaluating each candidate TE for a role in altering expression of proximal genes. In one approach, we determined whether polymorphic TEs could disrupt regulatory sequences, as annotated with the epigenomic data of the NIH Roadmap Epigenomics Consortium. In all, we identified 10 polymorphic TEs to examine further as causal candidates because they were positioned in enhancer, promoter, heterochromatin, or transcribed sequences present in neurologic tissues.
We hypothesized that the polymorphic TEs have a causal relationship with risk of psychiatric and neurologic disorders by altering expression of genes in cis. For evidence of altered gene expression, we queried the Genotype-Tissue Expression (GTEx) database, which contains expression data for 948 donors across 54 tissues. GTEx readily identifies changes in tissue-specific gene expression associated with loci-specific genetic variation. SNPs in LD are identified as eQTL (expression quantitative trait loci) if the genetic loci with the variants are significantly associated with altered expression of a gene in a specific tissue. We found that 31 of the TASs linked to TEs were variants that are associated with changes in expression of one or more adjacent genes within regions of the brain.
Having identified a number of polymorphic Alu elements that are significantly associated with disease risk detected by GWAS and that are correlated with altered gene expression in neurologic tissues by eQTL analysis, we developed a luciferase reporter assay to test whether the insert sequences in the context of flanking sequence can influence transcription activity. We measured the impact of candidate Alu and flanking sequences on the function of a minimal promoter in NCRM-1 (human neural stem cells). Of six candidate Alu insertions evaluated for their impact on promoter activity, we found that five significantly altered the expression of luciferase. Taken together, we identified 10 polymorphic TE insertions that are potential candidates on par with other variants for having a causal role in neurologic and psychiatric disorders.
Dense transposon integration reveals that essential cleavage and polyadenylation factors promote heterochromatin formation.
In eukaryotes, the assembly of DNA into highly condensed heterochromatin is critical for a broad range of functions related to genome integrity. The methylation of histone H3 on lysine 9 (H3K9me) is central to the formation of heterochromatin by creating binding sites for a range of chromatin proteins important for silencing transposable elements, chromosome segregation, and epigenetic inheritance. Used extensively for this purpose, S. pombe is an excellent model in which to study the molecular mechanisms that generate and regulate heterochromatin. Centromeres, subtelomeres, and the mating-type region are packaged into constitutive heterochromatin, while meiosis genes are silenced by facultative heterochromatin until cells are starved of nitrogen. Importantly, Clr4, the H3K9–specific histone methyltransferase, is recruited to heterochromatin regions by several mechanisms. Constitutive heterochromatin results from RNAi factors that include the Ago1 (a major component of RNA silencing complexes)–containing, RNA–induced transcriptional silencing complex (RITS). Facultative heterochromatin at meiosis genes is independent of RNAi and relies on the RNA elimination (i.e., degradation) of factors Red1 and Mmi1 and on the nuclear exosome. However, gaps exist in our understanding of how RNA elimination generates heterochromatin. A new approach to identifying gene function is the high-throughput sequencing of integration profiles, also known as Tn-Seq, which identifies genes important for growth under selective conditions. Genes necessary to sustain growth under a specific condition do not tolerate insertions in that condition. Tn-Seq has been applied to identify pathogenic genes in bacteria. However, we were the first to develop the method for a eukaryote [Reference 3].
With the goal of identifying novel factors important for heterochromatin, we produced dense profiles of integrations using the Hermes transposable element and a silencing reporter (ura4) positioned in the outer repeats of centromere 1. Inserts that disrupted genes important for heterochromatin activated ura4, and thus the cells were unable to grow when passaged in 5-fluoroorotic acid (FOA) (Figure 2A). Genes with established roles in heterochromatin assembly had significantly fewer insertions in cells with the centromere reporter otr1R::ura4 than in cells lacking the reporter (Figure 2B). The list of candidates consisted of a total of 199 genes and, importantly, 65 are known to be essential for viability. These essential genes were candidates because they tolerated many insertions in their 3′ sequences that reduced heterochromatin but not viability. The high number of essential genes is significant in that most proteins found to be important for heterochromatin are identified in screens of deletion strains that cannot include essential genes. The 199 candidates showed highly significant enrichments for functions in silencing at centromere outer repeats and included all four factors that produce siRNA.
Figure 2. Dense maps of transposable element integration identify genes important for heterochromatin at centromere repeats.
A. Single insertions of the transposable element Hermes were generated in cells with wild-type cen1 and cen1 otr1R::ura4. Cultures were passaged in 5-fluoroorotic acid (FOA) for 5 or 80 generations. Cells with insertions in heterochromatin genes (het1) express ura4 and cannot grow in FOA. After growth on FOA, fewer insertions were detected in het genes in cells with cen1 otr1R::ura4.
B. Genes involved in forming centromere heterochromatin such as mit1 and sir2 had fewer inserts in cells with the cen1 otr1R::ura4 (black, dupl. libraries) than in cells with WT cen1 (red, dupl. libraries).
We identified other RNA–processing factors that were not previously linked to heterochromatin structure. Strikingly, four of the RNA–processing candidates form an interaction module of the canonical mRNA polyadenylation and cleavage factor CPF, as predicted from highly homologous proteins in S. cerevisiae. To determine whether polyadenylation and cleavage contribute to heterochromatin structure at the centromere repeats, we focused on the function of Iss1, a subunit of CPF. We generated a C-terminal truncation of Iss1 (Iss1-deltaC) by removing 38 amino acids that, based on the Hermes insertions, were not important for viability. Iss1-deltaC showed no growth restriction on nonselective medium but exhibited a heterochromatin defect, as demonstrated by growth in the absence of uracil and reduced levels of H3K9 dimethylation (H3K9me2) at otr1R::ura4. The results demonstrated that the Hermes screen correctly identified Iss1 as important for heterochromatin structure at the otr1R::ura4 reporter. Interestingly, we found that Iss1 contributes to the heterochromatin of centromere repeats in cells that lack the otr1R::ura4 reporter but, in this case, the contribution to H3K9me2 was only observed when the RNAi pathway was disabled by deletion of ago1. This role at the outer centromere repeats is therefore independent or redundant with RNAi.
We expanded our study of the Iss1-deltaC mutation to evaluate changes in expression and transcription termination genome-wide. RNA-Seq data revealed that Iss1-deltaC did not significantly impact canonical transcription termination, but 73 genes were found to have higher expression. Importantly, the genes overlapped significantly with genes upregulated in cells lacking Rrp6, the 3′-5′ exonuclease subunit of the nuclear exosome. As a key subunit of the nuclear exosome, Rrp6 plays an important role in RNA surveillance in the degradation of meiotic transcripts expressed during vegetative growth and the resulting formation of heterochromatin at these genes. The elimination of meiotic mRNAs depends on the RNA–binding protein Mmi1 to bind to the determinant of selective removal (DSR) sequence in order to recruit the exosome. Our co-immunoprecipitation experiments revealed that Iss1 interacted with Rrp6, Mmi1, and the polyA polymerase Pla1, indicating that Iss1 is associated with this network of elimination factors. Significantly, the interaction with Mmi1 was disrupted by the Iss1-deltaC mutation, a mutation that greatly reduced H3K9me2 at meiotic genes. We tested whether Iss1 plays a direct role in the heterochromatin of meiotic genes by performing ChIP-Seq of Iss1-FLAG. While a subset of Iss1–bound genes were highly expressed and associated with the canonical function of Iss1 in mRNA termination, most Iss1–bound peaks showed a strong correlation with genes regulated by RNA elimination and heterochromatin. Importantly, the iss1-deltaC mutation caused significant increases in the RNA levels of these genes. Taken together, our studies of RNA levels, Iss1 association with chromatin, and H3K9me2 indicate that Iss1 plays a direct role in the formation of heterochromatin at meiotic genes. Our application of Hermes profiles to identify genes important for heterochromatin formation demonstrates the significance of the approach, especially given that we were able to identify large numbers of essential genes, a result not obtainable with other screens.
After these results were published, we received significant interest in the protocols we used for Tn-Seq with S. pombe. We compiled detailed instructions, provided specific timing, and described reagents [Reference 3]. The step-by-step method includes inducing transposition, selecting insertions that reduce heterochromatin, and generating libraries of insertions. Of particular note is an extensive discussion of troubleshooting together with suggested solutions.
Additional Funding
- FY2022 Office of AIDS Research Innovative Funds Program
Publications
- Li F, Lee M, Esnault C, Wendover K, Guo Y, Atkins P, Zaratiegui M, Levin HL. Identification of an integrase-independent pathway of retrotransposition. Sci Adv 2022 8:1–17.
- Ahn H, Worman Z, Lechsinska A, Payer L, Wang T, Malik N, Li W, Burns K, Nath A, Levin HL. Retrotransposon insertions associated with risk of neurologic and psychiatric diseases. EMBO Reports 2022 e55197:1–17.
- Li F, Hung S, Esnault C, Levin HL. A protocol for transposon insertion sequencing in Schizosaccharomyces pombe to identify factors that maintain heterochromatin. STAR Protoc 2021 2:100392.
Collaborators
- Kathleen Burns, MD, PhD, Dana-Farber Cancer Institute, Boston, MA
- Avindra Nath, MD, PhD, Division of Neuroimmunology & Neurovirology, NINDS, Bethesda, MD
- Mikel Zaratiegui, PhD, Rutgers, The State University of New Jersey, Piscataway, NJ
Contact
For more information, email henry_levin@nih.gov or visit https://sete.nichd.nih.gov.