Transcriptional Control of Cell Specification and Differentiation

- Jeffrey A. Farrell, PhD, Stadtman Investigator, Head, Unit on Cell Specification and Differentiation
- Paulina Capar, MSc, Technician
- Abhinav Sur, PhD, Postdoctoral Visiting Fellow
- Sean Lee, BS, Postbaccalaureate Intramural Research Training Award Fellow
- Avani Modak, BS, Postbaccalaureate Intramural Research Training Award Fellow
Animals consist of a collection of cells with diverse shapes, structures, and functions, a diversity that is rebuilt from scratch by every embryo. The genetic programs that direct the process are the central mystery of developmental and regenerative biology. We are interested in how decisions about the cell type to adopt are controlled, and how the genetic programs direct the morphological and functional specialization of different cells.
The single-cell revolution in developmental biology has given us new access and new tools to address these questions. I previously developed high-temporal-resolution, single-cell RNA–sequencing approaches to identify transcriptional trajectories, i.e., the ‘highways’ or the most likely paths through gene expression that cells take during development. From such data, we were able to identify the sequence of genes expressed by individual cell types during early development, which provides insight into the genetic programs that regulate cells’ choice of cell type and then their downstream functional transformations at a wider breadth than was previously achievable. Work in the lab focuses on more deeply exploring such processes, using the approaches we developed. Our lab combines single-cell genomics with imaging, genetic, and classical embryological approaches to investigate the genetic control of cell specification and differentiation during vertebrate embryogenesis. We focus on zebrafish embryos as a model system in which to study these questions, because, among vertebrates, they are easy to culture, image, and manipulate, both embryologically and genetically.
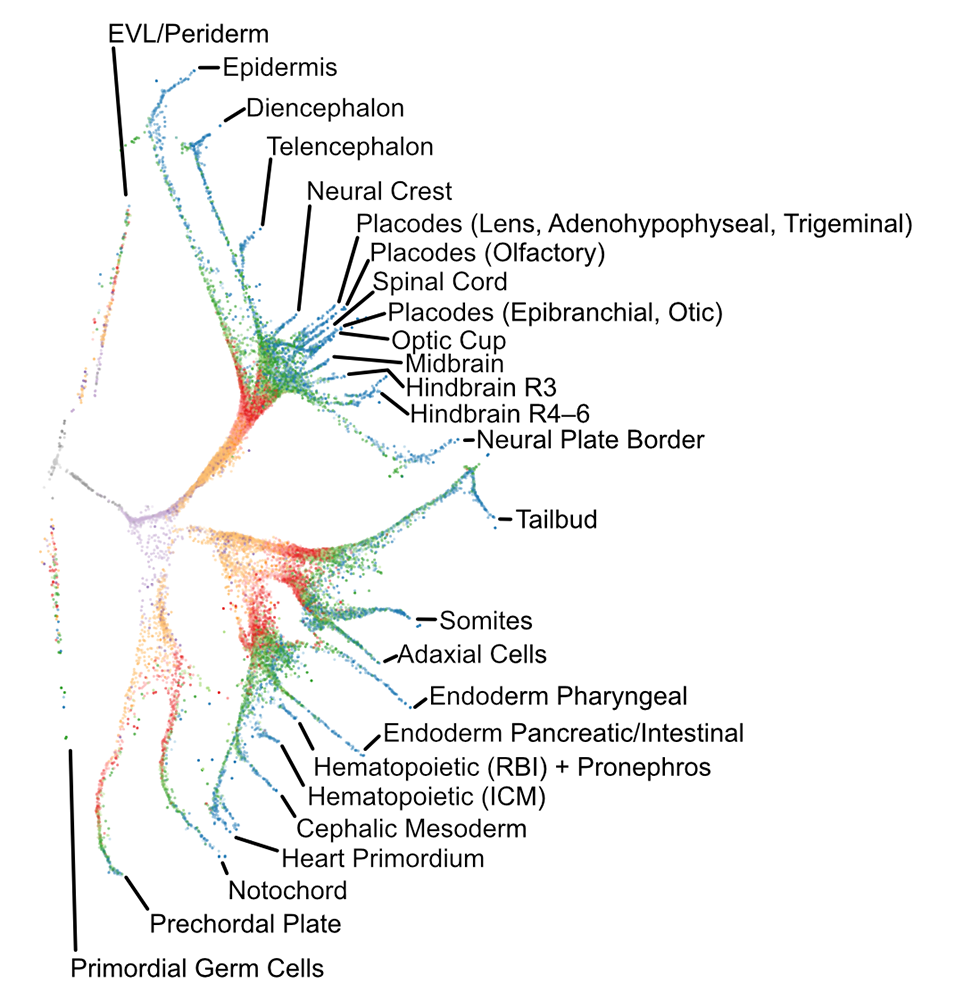
Figure 1. Transcriptional trajectories in early zebrafish development
Single-cell transcriptomes were isolated from zebrafish embryos at 12 developmental stages spanning 3–12 hours post-fertilization. The branching transcriptional trajectories, which represent the gene-expression events that give rise to 25 different differentiated cell types, were then reconstructed using URD, a simulated diffusion-based computational reconstruction method, which is software that we developed and published in 2018 [Reference 1].
Genetic underpinnings of cell differentiation
Once a cell has been specified, it must acquire the particular morphology and functionality of its cell type through the process of differentiation, a process that is driven by cell type–specific expression of differentiation genes and often results in dramatic changes in basic cell-biological processes. We aim to identify those genes that drive differentiation and understand their regulation. To do this, we generated a single-cell RNA-Seq atlas spanning embryogenesis and early larval stages (62 timepoints from 3–120 hours post-fertilization), annotated over 300 cell types, and built a web portal to enable other investigators to browse these data. To better understand how cells acquire their cell type–specific features, we also built a catalog of shared gene-expression programs that are re-used across many different tissues during development. Additionally, our analysis uncovered uncharacterized cell types in zebrafish development that have human homologs, which will be useful for building future disease models. We therefore built developmental trajectories to describe the sequence of gene expression that likely gives rise to these cell types, and we used these trajectories to determine candidate regulatory factors that may drive their specification and which can be tested in future work.
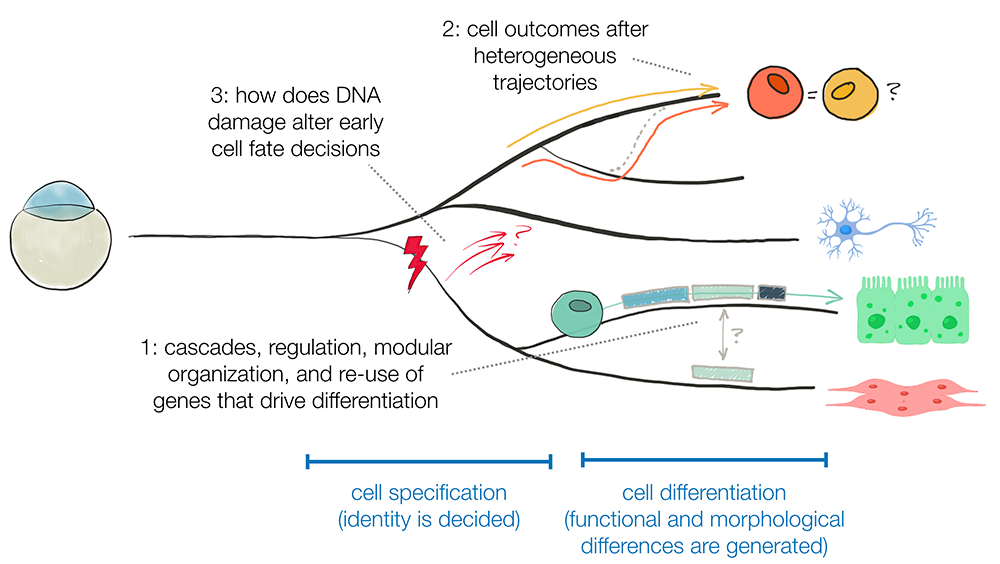
Consequences of heterogeneous developmental trajectories
Distinct cell types can arise through many developmental trajectories or developmental histories. We and others have observed refinement at the boundaries between groups of cells specified to become different tissues; at such boundaries, some cells switch from one specification state to another. We use the axial mesoderm as a model and seek to understand (1) what drives cell-type switching; (2) the long-term consequences for a cell that switched; and (3) the mechanisms that assist in successful switching.
Effect of environmental insults on developmental choices
During early embryogenesis, a field of equipotent cells are instructed to initiate different gene expression programs by external developmental signals and cell-intrinsic cues. We recently observed that cells that experience DNA damage in early zebrafish embryos initiate an unusual transcriptional response during a very limited window in development. Moreover, most damaged cells are not eliminated but appear to be excluded from contributing to some tissues in the animal, which suggests that responding to DNA damage may affect cells’ choices during development and which raises the question as to how that occurs. We are investigating (1) the fate of cells in early development that experience DNA damage; (2) the role this unusual transcriptional response plays; and (3) what drives the bias in damaged cells’ future cell type.
Publications
- Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, Schier AF. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 2018 360:eaar3131.
- Siebert S, Farrell JA, Cazet J, Abeykoon Y, Primack A, Schnitzler C, Juliano CE. Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science 2019 365:eaav9314.
- Raj B, Farrell JA, Liu J, El Kholtei J, Carte AN, Navajas Acedo J, Du LY, McKenna A, Relic D, Leslie JM, Schier AF. Emergence of neuronal diversity during vertebrate brain development. Neuron 2020 108:1058–1074.
Collaborators
- Celina Juliano, PhD, University of California, Davis, Davis, CA
- Michal Rabani, PhD, The Alexander Silberman Institute of Life Science, The Hebrew University of Jerusalem, Jerusalem, Israel
Contact
For more information, email jeffrey.farrell@nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/farrell.