Molecules and Therapies for Craniofacial and Dental Disorders
- Rena D’Souza, MS, DDS, PhD, Head, Section on Craniofacial Genetic Disorders, Director of the National Institute of Dental and Craniofacial Research
- Fahad K. Kidwai, BDS, MSc, PhD, Clinical Staff
- Resmi Raju, PhD, Postdoctoral Fellow
- Parna Chatteraj, Lab Manager
- Jeremie Oliver Piña, MS, MBA, Predoctoral Fellow
Embryonic development of the craniofacial complex requires tightly controlled molecular crosstalk between proliferating and differentiating cell networks to form the most intricate structures in the human body. During embryogenesis, perturbations in environmental and/or genetic milieu can negatively affect craniofacial development. While considerable progress has been made in studying isolated genetic mutations leading to syndromic and non-syndromic craniofacial disorders, the broad molecular-genetic mechanisms driving morphogenetic events have yet to be sufficiently explored and understood. Such gaps in our understanding have restricted clinical treatment options for patients affected by common developmental anomalies, such as cleft palate or tooth agenesis. Thus, there is a strong biologic rationale for a thorough investigation of basic molecular mechanisms driving craniofacial structure morphogenesis, which may pave the way toward translatable therapeutic developments for patients.
The overarching goal of our research program is to conduct basic and translational studies of genetic and molecular mechanisms involved in craniofacial development, with the primary aim of unveiling novel regulatory molecules and putative patient-centric therapeutic solutions for craniofacial and dental disorders. Among the molecular pathways known to control craniofacial development is Wnt/β-catenin signaling. Wnts are also well known as upstream effectors of osteogenesis and odontogenesis. Our lab demonstrated, for the first time, the successful in utero correction of cleft palate defects in a Pax9–/– mouse genetic model by small-molecule neutralizing therapy targeting Wnt–antagonizing proteins. We are now actively investigating additional drug-delivery approaches and molecular targets for the modulation of Wnt signaling in vivo to permit targeted correction of both cleft palate and tooth agenesis. Our research group employs basic principles of developmental biology, next-generation sequencing, regenerative medicine, tissue engineering, and drug delivery models to identify and validate novel approaches to restore molecular equilibrium in genetic models of the highly relevant human diseases cleft palate and tooth agenesis. Actively fostering collaborations with both intramural and extramural investigators, from basic scientists, engineers, to clinicians, the lab aims to pioneer innovative approaches toward the treatment of patients affected by craniofacial disorders of development. Through these rigorous and robust preclinical, proof-of-principle studies, we hope to open up pathways toward novel clinical trials for the treatment of previously unpreventable disorders affecting the craniofacial complex.
Toward a novel in utero therapy for the prenatal cure of cleft palate in a mouse genetic model
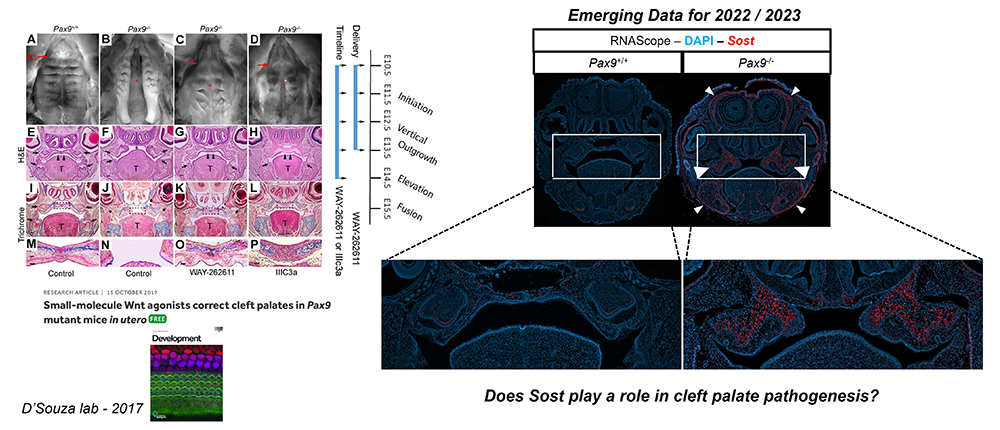
Cleft palate (CP), together with cleft lip, is among the most common birth defects in humans, occurring in up to 1 in 500 live births. Such birth defects can inflict heavy physical, mental, psychosocial, and financial burdens on patients and their caregivers throughout life, often requiring many stages of complex surgical correction with varying success rates. Hence, there remains a substantial need for innovative approaches to alleviate the burden of postnatal care for these patients. While considerable progress has been made in studying isolated genetic mutations leading to syndromic and non-syndromic cleft disorders, the broad molecular genetic mechanisms driving osteogenic differentiation from palatal shelf out-growth, elevation, and fusion events have yet to be sufficiently explored and understood. Such gaps in our understanding have restricted clinical treatment options for patients affected by cleft disorders. Given that prenatal molecular diagnostics have made CP identifiable earlier in gestation, the advancement of safe and efficacious interventions in utero to correct CP has become feasible.
The objective of our work is to investigate the molecular genetic mechanisms driving palatal osteogenesis and to optimally manipulate key signaling environments in utero to promote CP correction in a mouse genetic model. If properly explored, such information can be applied to the development of novel therapeutics that can benefit individuals with isolated or syndromic CP defects who face complex surgeries and the arduous burden of life-long care. The long-term goal of this research is to investigate the spatiotemporal molecular mechanisms driving osteogenic differentiation in normal palate development and in CP dysmorphogenesis, using Pax9–/– as a model. Differential multiomic profiles of expression in spatial biological context will unveil the molecular framework for the development of novel therapeutic strategies to optimize signaling environments during development. The proposed research will test the following hypotheses: (1) Wnt signaling effector function is critical for osteogenesis of the embryonic palate; and (2) loss of the up-stream master regulator of Wnt signaling homeostasis, Pax9, results in the disruption of palatal osteogenesis via up-regulation of sclerostin (Sost), a potent inhibitor of Wnt signaling and bone formation (Figures 1&2). The specific aims of this proposal are as follows: (1) to define the spatiotemporal transcriptomic profile of embryonic palate osteogenesis via unbiased stage-specific signature mapping of cell populations in the normal murine secondary palate; (2) to differentially compare epigenomic, proteomic and gene expression signatures of Wnt–related osteogenesis in Pax9-/- CP; and (3) to pilot novel in utero drug delivery approaches and molecules, based on the molecular profiles observed in situ, for the prenatal cure of CP. The proposed research will add novel foundational knowledge of multiomic morphogenetic expression gradients of key Wnt signaling regulators within the embryonic palate and will propose an innovative therapeutic model whereby palatal clefts may be corrected in utero.
The proposed research will improve unbiased mechanistic understanding of palatal bone development and carry out the first preclinical study of intra-amniotic small-molecule and antibody-replacement drug delivery for targeted palatal osteogenesis. The validation of a translational in utero drug delivery system for reversal of single-gene CP disorders will lead to preventive and corrective prenatal therapeutic interventions in humans.
Role of Wnt signaling pathway genes in dentinogenesis
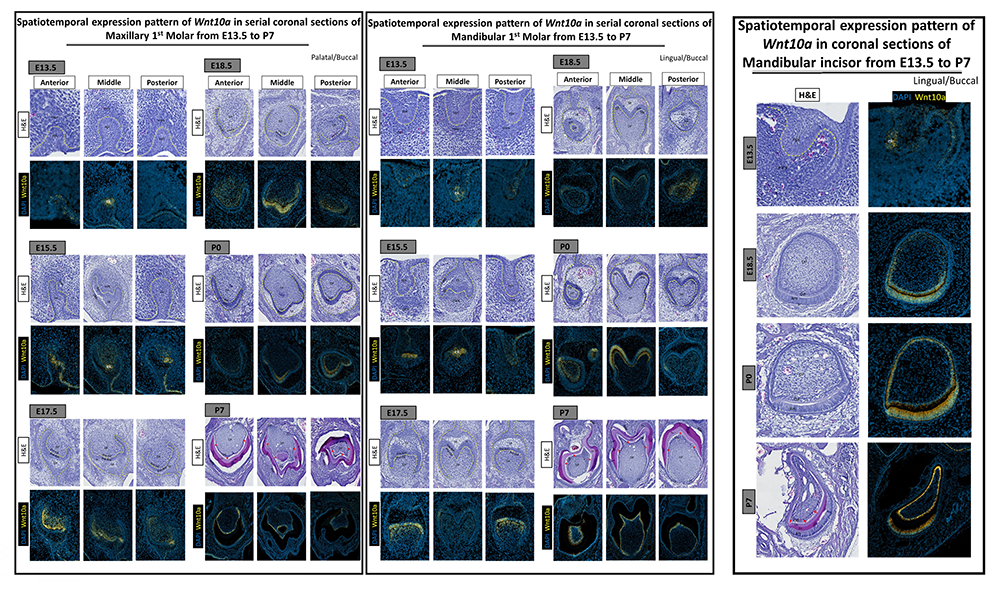
Dental caries is one of the most common diseases worldwide. Two mineralized tissue barriers in the tooth are composed of an outer and inner layer of enamel and dentin, respectively. Trauma or dental caries can lead to the destruction of this mineralized barrier as well as inner specialized cell layer (odontoblast cells) in the dental pulp. The odontoblast cell layer is responsible for forming a dentin bridge (reparative dentinogenesis) to protect the inner vital pulp tissue. However, the process of reparative dentinogenesis is limited to the critical size of the tooth caries. In a large dental decay, the inner pulp is exposed to the oral cavity and, if not treated at an early stage, can undergo necrosis. Traditional clinical treatments used to replace the affected tooth structure are based on artificial filling materials. This highlights the importance of developing a therapy that stimulates and directs the cells in dental pulp to repair injuries afflicting dentin and pulp. One of the main focuses of regenerative dental research is to preserve the natural architecture of tooth structure following deep dental cavities. Modern regenerative dentistry challenges us to apply the current scientific knowledge of developmental biology into translational dental medicine. The Wnt/β-catenin signaling pathway plays a major role in dentinogenesis, as well as dentin repair following tooth damage. Wnt/β–catenin signaling has thus emerged as a major target in dentin regeneration and repair by overstimulating it in a variety of ways (Figures 3&4). However, there is a gap in our understanding of how modulation of the Wnt/β–catenin signaling genes that are involved during dentinogenesis and reparative dentinogenesis occurs in a time- and region-specific manner.
Our preliminary data confirm the spatiotemporal involvement of Wnt10a in odontoblast differentiation and dentinogenesis. A synergistic expression pattern of Wnt10a and the Wnt regulators Sost and Dkk1 was observed in early stages of normal embryonic tooth development. At the later stage of P7, following enamel and dentin development, Wnt10a expression is confined to a single layer of odontoblasts and reduced dental epithelium with a strong expression in the cervical loop area. Expression of the Wnt modulators Sost and Dkk1 was reduced or disappeared at P7 developmental stage. Gradient expression pattern of Wnt10a expression in ameloblast and odontoblast along the cusp tips and grooves suggests the involvement in cusp morphogenesis.
Identifying and comparing the spatiotemporal expression of Wnt signaling molecules in dentin development and the dentin repair process will provide new insight into the nature of dentin regeneration and help us identify potential targets for regenerative therapy in dentistry.
Transcriptomic analysis reveals that differential signaling events control maxillary and mandibular incisor development.
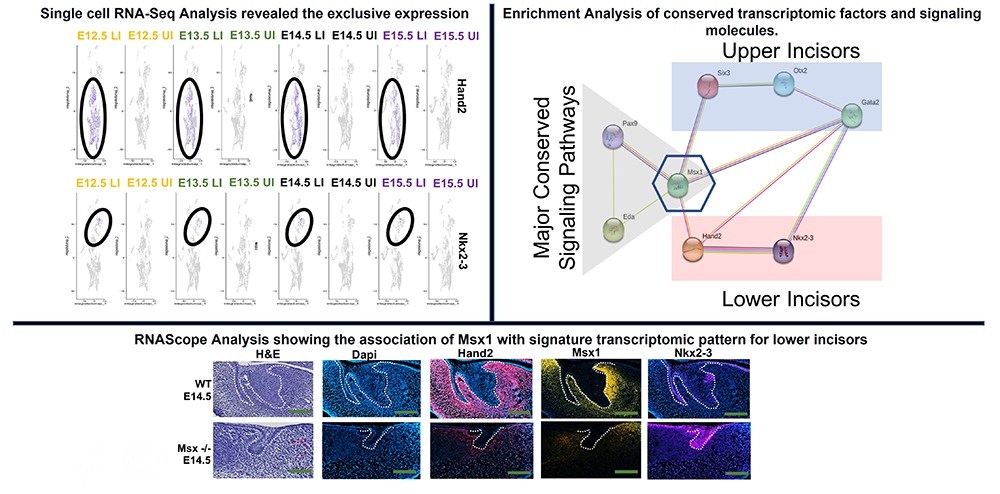
Tooth development is a complex process involving the reciprocal interactions between the dental epithelium and the underlying mesenchyme. Morphogenic gradients mediate this epithelium-mesenchymal interaction. It is known that morphogenic gradients influence the distinct patterning of dentition. The broad range of patterning in dentition provides a valuable system in which to study the heterogeneity in molecular mechanisms during the process of odontogenesis. However, little is known about the distinct morphogenic gradients or signature transcriptomic factors responsible for the formation of mouse incisiform in the maxillary vs. mandibular domain. Our study therefore focuses on identifying the exclusive morphogenic gradients or signature transcriptomic factors within the maxillary vs. mandibular incisiform field during the early odontogenesis.
Our unbiased next-generation sequencing experiments revealed exclusive morphogenic gradients and signature transcriptomic patterns expression in the maxillary (Six3, Otx2 and Garta2) vs mandibular incisiform (Hand2 and Nkx2-3) field. Our Enrichment Analysis revealed Msx1 as a master regulator of signature transcriptomic patterns of maxillary and mandibular incisors (Figure 6). Our preliminary data show an association of Msx1 with Hand2 in lower incisiform field (Figures 5&6). Msx1 deficiency also results in sporadic expression of Nkx2-3 unlike in wild-type, where the expression was found to be confined to enamel knot (Figures 5&6). Experiments to identify the association of Msx1 with Six3, Otx2 and Gata2 in maxillary incisiform development are under way.
For the first time, these studies provide a rationale for the differential effect of distinct morphogenetic gradients or signature transcription factors regulating incisiform in the maxillary vs. mandibular domain. The findings provide a framework for future studies to understand the patterns of tooth agenesis. Patterns of tooth agenesis provide valuable clues about the important roles of exclusive transcription factors and distinct morphogenic gradients in modulating during incisiform development in the maxillary vs. mandibular domain.
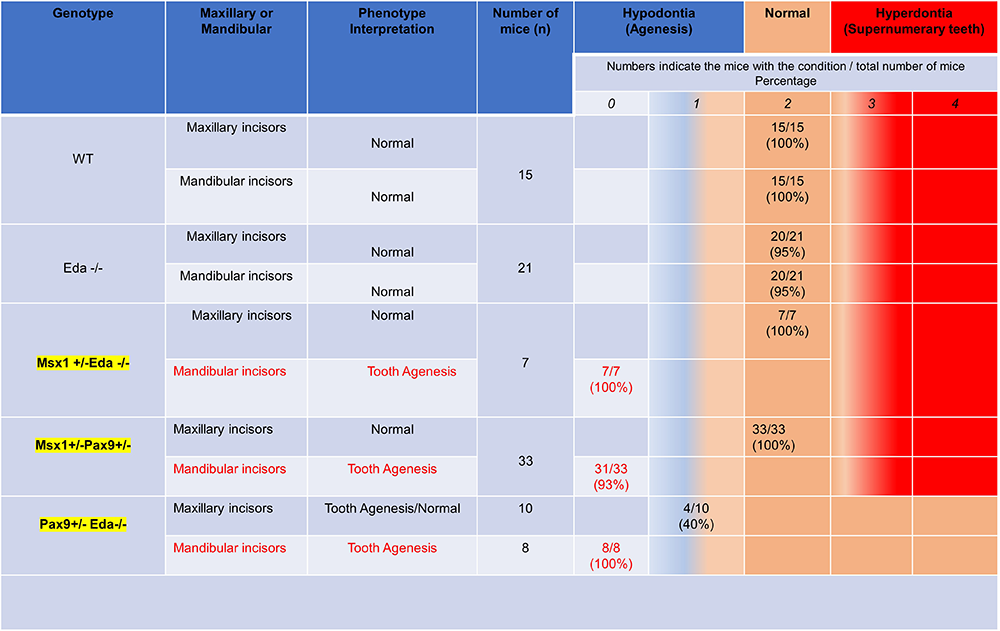
In vivo molecular consequences of Wise deficiency on Pax9 and/or Msx1–dependent signaling events that control maxillary vs mandibular incisors
It is known that morphogenic gradients influence the patterning of dentition, and we and others have shown that transcription factors such as Msx1 and Pax9 modulate the functions of Bmp, Wnt, and Eda during odontogenesis (Figure 7). Recently, inhibition of a known bifunctional BMP and Wnt antagonist, Wnt modulator in surface ectoderm (Wise), was shown to selectively rescue tooth formation in mouse genetic models. Whether Msx1 interacts with Wise and Eda in the incisor domain remains unclear. The goal of this study was to analyze the in vivo molecular consequences of WISE deficiency on Msx1–dependent signaling events that control maxillary vs mandibular incisors. We successfully generated the disease model with Msx1, Pax9 and Eda strain. Our preliminary data revealed a unique pattern of tooth agenesis (lower incisors).
For the first time, our studies provide a rationale for the differential effects of the Bmp and Wnt pathways in regulating Msx1–dependent events that pattern the distal maxilla and mandibular domains. The data provide a framework for the development of targeted therapeutics for human tooth agenesis.
Figure 7.
Disease model for tooth agenesis
Download this table as a PDF: dsouza_2022_fig7.pdf (PDF, 68 KB)
Publications
- Piña JO, Raju R, Chattara P, Kidwai FK, D’Souza RN. Spatiotemporal mapping of the embryonic palate transcriptome. Commun Biol 2022; under revision.
Collaborators
- Fabio R. Faucz, PhD, Molecular Genomics Core, NICHD, Rockville, MD
- Steven L. Goudy, MD, MBA, FACS, Emory University School of Medicine, Atlanta, GA
Contact
For more information, email rena.d'souza@nih.gov.