The Biophysics of Protein-Lipid Interactions in Influenza and Coronavirus, Malaria, and Muscular Dystrophy

- Joshua Zimmerberg, MD, PhD, Head, Section on Integrative Biophysics
- Paul S. Blank, PhD, Staff Scientist
- Svetlana Glushakova, MD, PhD, Staff Scientist
- Matthias Garten, PhD, Visiting Fellow
- Irene Jimenez Munguia, PhD, Visiting Fellow
- Yuto Kegawa, PhD, Visiting Fellow
- Hang Waters, MS, Biologist
- Jennifer Petersen, PhD, Electron Microscopist
- Elena Mekhedov, MA, Contractor
- Tatyana I. Tenkova-Heuser, PhD, Contractor
- John E. Heuser, MD, Senior Biophysicist
- Glen Humphrey, PhD, Guest Researcher
- Adriana Golding, PhD, Postdoctoral Intramural Research Training Award Fellow
- Garrett Tisdale, BA, Postbaccalaureate Trainee
- Jaqulin Wallace, MS, Postbaccalaureate Trainee
Eukaryotic life must create the many shapes and sizes of the system of internal membranes and organelles that inhabit the variety of cells in nature, membranes that must remodel for cells to repair damaged plasmalemma and deal with infectious agents such as viruses and parasites. Such basic membrane mechanisms must be highly regulated and highly organized in various hierarchies in space and time to allow the organism to thrive despite environmental challenges, genetic instability, unpredictable food supply, and physical trauma. We are using our expertise and the techniques we perfected over the years to address various biological problems that have in common the underlying regulation or disturbance of protein/lipid interactions. The overall goal of this project is to determine the physico-chemical mechanisms of membrane remodeling in cells. This year, we focused on enveloped viral assembly and cell entry, and the biology of extracellular vesicles.
Delta variant SARS-CoV-2 spike protein causes viral aggregation
Genetic variants of the severe acute respiratory syndrome coronavirus 2, SARS-CoV-2, continue to evolve as the virus circulates worldwide, and each variant holds the potential to evade acquired immunity and re-ignite the COVID-19 pandemic. SARS-CoV-2 is an enveloped RNA virus containing a single-stranded, positive-sense genome. Prominent, club-shaped spike glycoproteins (spikes) project from the viral envelope, mediating binding and fusion between the viral envelope and host-cell membranes to deliver the viral genome. Spikes are highly immunogenic, eliciting a robust neutralizing antibody response; the immunogen is encoded by the extremely efficacious mRNA vaccines. Given its essential role in viral entry and immunity, mutations occurring on the spike require careful genetic, structural, and functional surveillance.
The fully assembled, prefusion spike consists of a trimer of spike protomers, each of which is highly glycosylated. Proteolytic cleavage of the SARS-CoV-2 spike by furin during biosynthesis nicks the spike into two subunits, S1 and S2. The S1 subunit contains the receptor-binding domain (RBD), which can be in an up (receptor accessible) or down (receptor inaccessible) conformation, as well as the N-terminal domain (NTD) and two C-termini. S1 caps the S2 subunit, which harbors the membrane-fusion machinery. The spike is highly flexible owing to a hinged stalk, helping the RBD bind to angiotensin converting enzyme II (ACE2), the host-cell receptor. Once attached to the host cell by ACE2 binding, a second cleavage event occurs in S2 by a host-cell protease, either TMPRSS2 at the cell surface or cathepsin in the endosomal membrane, depending on cell type, protease availability, and viral variant. Upon S2 cleavage, the fusion peptide undergoes large conformational changes, which drive fusion between membranes.
Millions of SARS-CoV-2 genomic sequences have been cataloged since the original strain emerged in Wuhan, China, in December 2019. Amino acid substitutions or deletions in the spike that impart fitness benefits to the virus have evolved and sometimes converged independently in different geographical locations, giving rise to several variants designated Variants of Concern (VOCs). By May 2020, the D614G variant, designated Pango lineage B.1, rapidly supplanted the original Wuhan strain globally, and the D614G substitution is present in all subsequent variants. Next, the VOC Alpha (lineage B.1.1.7) emerged in the UK and became dominant worldwide by early 2021.
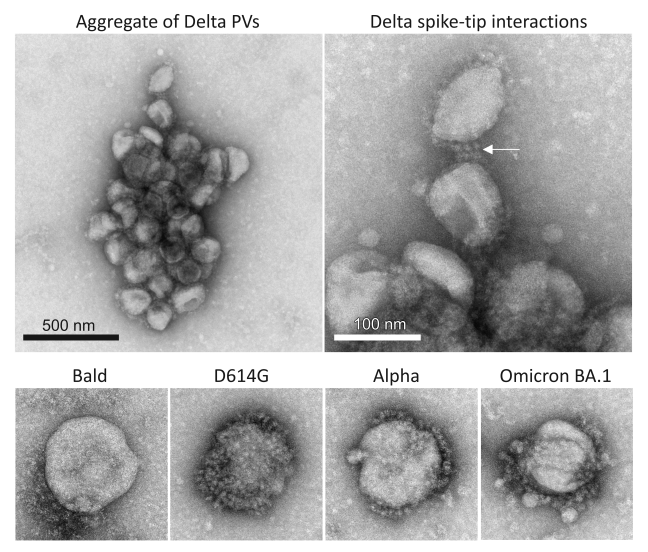
The Alpha variant prevailed until it was outcompeted by VOC Delta, a member of the B.1.617 lineage, which arose in India and became globally dominant in mid-2021. The Delta variant was considerably more transmissible than Alpha, infected individuals faster, produced earlier detection by PCR test with higher viral load, and was more pathogenic. The Delta variant's spike displayed a different combination of mutations than Alpha. Functional consequences of spike mutations can be tested using engineered mimics of enveloped viruses, called pseudotyped viral particles (PVs). PVs are produced by co-transfecting producer cells with plasmids encoding a capsid protein from a parental virus (typically a retrovirus or arbovirus capsid), the spike glycoprotein of interest, and a reporter gene that produces a fluorescent or luminescent protein signal upon host cell entry. The capsid core buds efficiently from the producer cell while incorporating the heterologous spike and encapsulating the reporter gene. To search for an ultrastructural correlate to the increased initial infection rate of Delta PVs, we took advantage of our recent experience with PVs packaged with various SARS-CoV-2 spike variants into target cells, reported last year. PVs bearing the Delta spike (Delta PVs) drove markedly faster initial infection, and greater infection overall, than other variants, a phenotype shared by Delta SARS-CoV-2. Using four independent techniques (negative stain TEM, flow cytometry, NTA, and cryo-EM), we demonstrated that PVs bearing the Delta variant spike or the closely related Delta sublineage Delta AY.4.2 spike aggregate, whereas PVs bearing other spike variants and Bald PVs do not aggregate. Given that the PVs were prepared in parallel, handled under identical conditions, and factors that could promote aggregation such as pH changes, freeze-thaw cycles, and high-speed ultracentrifugation were avoided, the observed aggregation of Delta PVs most likely reflects a unique property of the Delta and Delta AY.4.2 spikes. The observation that Delta and Delta AY.4.2 PVs continued to aggregate in solution while stored at 4°C suggests that aggregation occurs after budding from the producer cell; however, interaction between PVs could also initiate during biosynthesis and budding from the producer cell [Reference 2].
Unlike SARS-CoV-2, which buds into the ER–Golgi intermediate compartment (ERGIC) during final assembly, retroviruses, including MLV, generally bud directly from the plasma membrane (but not always). Thus, it is not known when or whether Delta variant SARS-CoV-2 aggregates, and this is currently being investigated. Delta SARS-CoV-2 could aggregate while budding into the ERGIC, during egress through alkalized lysosomal organelles, after egress in the extracellular milieu, or even on the target-cell plasma membrane. The ability to aggregate may depend on the concentration of viral particles in each environment. The fact that Delta PVs continue to aggregate while stored at 4°C is consistent with a mass action mechanism for Delta PV aggregation. Measurements of the rate of aggregation of Delta PVs at temperatures ranging up to physiological temperature could shed light on the thermodynamic properties of Delta aggregation, and advance understanding of the mechanism of aggregation. Furthermore, to produce PVs, a 19 amino-acid C-terminal truncated version of each variant spike was expressed, which has been shown to increase the amount of spike incorporated into the PV envelope and raise PV infectivity. Truncations in the cytoplasmic tail could modify properties of the spike ectodomain structure and function. Although each variant spike possessed the same truncation, it is not impossible that the truncation uniquely affected the Delta spike ectodomain, conferring the aggregation property.
Examples of Delta PVs with apparent spike tip interactions were observed by negative staining, but these interactions did not extend to lateral aggregation of spike proteins on the surface of a PV. Ongoing cryo-electron tomography studies will reveal the nature of the interactions between aggregated PVs. Spike-mediated aggregation differs from antibody-driven aggregation of virions expected from polyvalent neutralizing antibodies, as those binding constants are expected to be much stronger. Thus, virions would be tightly packed and not likely to disaggregate at the cell surface. The spike tip interactions are more likely to come apart upon surface binding and receptor competition for the RBD at the tip of the spike.
Analysis of the mutations present in the Delta and Delta AY.4.2 spikes compared with other variants may provide clues as to their unique property to aggregate. Delta and Delta AY.4.2 amino-acid sequences are similar, except for four additional substitutions in Delta AY.4.2 (T95I, Y145H, A222V, and K458R). T95I is also present on the Omicron BA.1 spike, and residue Y145 is also substituted in Omicron BA.1 (Y145D). A222V and K458R are unique to Delta AY.4.2. Given that the presence of these mutations in Delta AY.4.2 does not abrogate or enhance the aggregation of Delta AY.4.2 compared with Delta, they appear to have no effect on aggregation.
Most of the other mutations on the Delta and Delta AY.4.2 spikes are shared with other variants. The D614G mutation is present in all variants, the substitution L452R, located in the RBD, is present in the Kappa variant (B.1.617.1), and a similar substitution L452Q occurs in the Lambda variant (C.37). A second RBD substitution at T478K is also found in Omicron BA.1 and BA.2. The mutation P681R is present in Kappa, and P681H exists in Alpha, Omicron BA.1, BA.2, and Mu (B.1.621). Substitution mutation D950N is also shared with Mu. Because the non-Delta variants studied here do not aggregate, it is unlikely that any of their Delta-shared mutations can be aggregation-dominant. There are, however, three residues, E156, F157, and R158, in the NTD of Delta and Delta AY.4.2 that are uniquely and identically mutated: substitution E156G, and deletions at F157 and R158. It is possible that these three mutations in the NTD are sufficient to bestow the aggregation property alone or in the context of the other Delta mutations.
The clustering of Delta PVs could account for the faster and larger initial infection observed in entry assays with the Delta PVs. Because the number of spike trimers is larger on an aggregate comprising multiple PVs, and the cell-surface contact area is larger for any collision between the aggregate and a target cell, the effective on-rate for aggregate binding should be larger, resulting in faster binding. Furthermore, the avidity of the aggregate for the target cell would be enhanced manyfold owing to the many potential binding partners on a single contacting surface. Moreover, the increased dwell time at that contact area will allow for diffusional and conformational motions of proteins and lipids to increase the chance of membrane fusion, as these factors are important for avoiding hemifusion and promoting full fusion. All these factors should lead to the relatively higher initial rate of PV entry into target cells from aggregated PVs. Whether or not aggregates could enable the simultaneous delivery of multiple copies of entry reporter genes to target cells is not clear, given that the PVs need not display ACE2 and may thus not fuse with each other, even in an endosome; thus, each virus in an aggregate may have to independently fuse with the endosomal membrane to place its genes to that cell's cytoplasm. Implicitly, there would be more overall binding events for unaggregated PVs, each at another site. However, if the probability of PV entry was low owing to unbinding, then the factors to increase PV avidity, discussed above, would tend to increase overall fusion and its rate.
In summary, an ultrastructural analysis of retrovirus pseudotyped viral particles bearing SARS-CoV-2 spike variants led to a serendipitous discovery of significant aggregation when the Delta variant spike was expressed, but not upon expression of three other variant spikes [Reference 2]. Viral aggregation can impart fitness benefits by protecting virions from environmental hazards and by effecting simultaneous delivery of multiple viral genomes, or collective infection. Notably, collective infection can favor initial infection in some contexts. Likely, the size of an aggregate, and therefore the number of virions per aggregate, is important. If an aggregate is too large, subsuming many virions, it would effectively reduce infectious units below a threshold. On the other hand, if there are too few virions in an aggregate, the benefit of collective infection is not gained. The unique property of the Delta spike to aggregate PVs may underlie the faster infection seen by Delta PVs. Furthermore, spike-mediated aggregation could be part of the molecular mechanism by which Delta variant SARS-CoV-2 achieves increased transmissibility and faster infection with a higher viral load. The continued aggregation of PVs over time indicates that clustering may be mediated by interactions between spike tips, which in turn may indicate an adhesivity of the viral surface recognized by the immune system, thus altering the balance of host antiviral response towards inflammation.
Control of RNA packaging into extracellular vesicles by CD47
Extracellular vesicles (EVs) are vesicles released by all cells that are loaded with specific cargoes, and they mediate intercellular communication in physiological and disease states. CD47 is a signaling receptor for thrombospondin-1 (TSP1), the secreted extracellular matrix protein, which regulates a variety of cellular responses. CD47 is a component of extracellular vesicles (EVs) released by various cell types, including T cells. CD47 loads EVs with specific subsets of coding and non-coding RNAs that impart functional effects on target cells, including endothelial cells. How CD47 controls the loading of specific RNAs into EVs is unknown. Previous studies showed that EVs produced by CD47–knockout (KO) T cells lost the ability to modulate gene expression and signaling in recipient T cells and vascular endothelial cells, suggesting that the packaging of RNAs into specific subpopulations of EVs is regulated by CD47.
We examined the mechanisms by which CD47 directly or indirectly regulates which RNAs are packaged into T cell EVs. Mass spectrometry, biochemistry, light and electron microscopy, pharmacological manipulation, and genetic analyses were used to parse the mechanism by which CD47 controls the loading of EVs with specific RNAs. It was demonstrated that CD47 regulates nuclear/cytoplasmic transport of m7G–capped RNAS and their abundance in EVs via a physical interaction of CD47 and its cytoplasmic signaling adapter ubiquilin-1 with exportin-1 and several regulators of its nuclear export complex. The CD47/ubiquilin-1 complex regulates intracellular trafficking of capped miRNAs and mRNAs and their trafficking into EVs. These results establish TSP1/CD47 signaling as a regulator of nuclear/cytoplasmic RNA trafficking and the subsequent packaging and release of a subset of 5-7-methylguanosine-modified (m7G) RNAs in EVs. The relevance of m7G-cap–dependent RNA trafficking to the physiological functions of CD47 in cardiovascular disease, aging, cancer, and infection remain to be investigated [Reference 5].
Regulation of umbilical cord endothelial cells by extracellular vesicles
During pregnancy, umbilical cord endothelial cells engage in bidirectional communication that regulates angiogenesis. EVs play important roles in cell-cell communication by transferring bioactive molecules, including mRNAs and miRNAs, into recipient cells. EVs are taken up by recipient cells, where they modulate the fate of those cells, including altering gene expression. EVs produced by a given cell type are heterogeneous in their RNA content, but it is unclear how specific EV surface markers relate to their functional effects on target cells. Our previous work established that EVs bearing CD63, MHC-I, or CD47 surface markers contain distinct noncoding RNA populations. Thus, when applied to endothelial cells, EVs produced by wild-type (WT) T cells (that express CD47) compared with mutant T cells lacking CD47, lead to differential modulation of VEGF–induced cell proliferation, tube formation, and VEGFR2 phosphorylation, findings that reflect the divergent RNA compositions of the EVs produced by these different cells, leading to distinct functions in target cells [Reference 6].
In this project, we compared the effects on Human Umbilical Vein Endothelial cell (HUVEC) gene expression of CD63+ and MHC-I+ EV subsets derived from WT T cells and CD47-KO T cells. The goal was to identify several functional gene families in HUVECs that are differentially regulated by the two EV populations. The present study uses biochemical approaches, genetic sequencing, and electron microscopy to evaluate the EVs isolated from the cell types. The study reveals that CD63+ and MHC-I+ EVs from CD47-KO T cells are enriched in small non-coding RNAs compared with EVs from WT T cells. CD47–deficient T cells secrete more CD63+ and MHC-I+ EVs, but MHC-I+ EVs are selectively taken up more by HUVECs. Transcriptomics analysis of endothelial cells treated with CD63+ or MHC-I+ EVs showed surface marker– and CD47–dependent changes in gene expression in the target cells. Gene-set enrichment analysis identified effects of T–cell EVs on VEGF and inflammatory signaling, cell cycle, and lipid and cholesterol metabolism. Thus, subsets of T–cell EVs differentially regulate endothelial cell metabolism and inflammatory and angiogenic responses. Future studies will focus on identifying mechanisms by which CD63 and MHC-I regulate EV uptake and the distinct functional effects of MHC-I+ EVs and CD63+ EVs on endothelial cells and other types of target cells. These studies may identify additional functions of EVs produced by T cells that regulate physiological angiogenic and inflammatory pathways and antitumor immunity.
Figure 1. SARS-CoV-2 virus entry particles
Negative stain electron microscopy images of PVs bearing various SARS-CoV-2 spike proteins show that PVs bearing Delta spikes assemble into unique aggregates compared with PVs bearing other spike variants (D614G, Alpha, Omicron BA.1) or PVs bearing no spike protein (Bald), which exist mainly as single PVs. Interactions between the tips of Delta spike proteins may mediate Delta-specific aggregation.
Publications
- Jiménez-Munguía I, Beaven AH, Blank PS, Sodt AJ, Zimmerberg J. Interferon-induced transmembrane protein 3 (IFITM3) and its antiviral activity. Curr Opin Struct Biol 2022 77:102467.
- Petersen JD, Lu J, Fitzgerald W, Zhou F, Blank PS, Matthies D, Zimmerberg J. Unique aggregation of retroviral particles pseudotyped with the delta variant SARS-CoV-2 spike protein. Viruses 2022 14:1024.
- Xu M, Pradhan M, Gorshkov K, Petersen JD, Shen M, Guo H, Zhu W, Klumpp-Thomas C, Michael S, Itkin M, Itkin Z, Straus MR, Zimmerberg J, Zheng W, Whittaker GR, Chen CZ. A high throughput screening assay for inhibitors of SARS-CoV-2 pseudotyped particle entry. SLAS Discov 2022 27:86–94.
- Pfeiffer A, D Petersen JD, Falduto GH, Anderson DE, Zimmerberg J, Metcalfe DD, Olivera A. Selective immunocapture reveals neoplastic human mast cells secrete distinct microvesicle- and exosome-like populations of KIT-containing extracellular vesicles. J Extracellular Vesicles 2022 11:e12272.
- Kaur S, Saldana AC, Elkahloun AG, Petersen JD, Arakelyan A, Singh SP, Jenkins LM, Kuo B, Reginauld B, Jordan DG, Tran AD, Wu W, Zimmerberg J, Margolis L, Roberts DD. CD47 interactions with exportin-1 limit the targeting of m7G-modified RNAs to extracellular vesicles. J Cell Commun Signal 2022 16:397–419.
- Kaur S, Elkahloun AG, Petersen JD, Arakelyan A, Livak F, Singh SP, Margolis L, Zimmerberg J, Roberts DD. CD63+ and MHC Class I+ subsets of extracellular vesicles produced by wild-type and CD47-deficient Jurkat T cells have divergent functional effects on endothelial cell gene expression. Biomedicines 2021 9:1705.
Collaborators
- Sergei Akimov, PhD, Frumkin Institute of Physical Chemistry and Electrochemistry, Russian Academy of Sciences, Moscow, Russia
- Oleg Batishchev, PhD, Frumkin Institute of Physical Chemistry and Electrochemistry, Russian Academy of Sciences, Moscow, Russia
- Andrew Beaven, PhD, Unit on Membrane Chemical Physics, NICHD, Bethesda, MD
- Christopher Karl Ernst Bleck, PhD, Electron Microscopy Core Facility, NHLBI, Bethesda, MD
- Wendy Fitzgerald, BA, Section on Intercellular Interactions, NICHD, Bethesda, MD
- Vadim Frolov, PhD, Universidad del País Vasco, Bilbao, Spain
- Timur Galimzyanov, PhD, Frumkin Institute of Physical Chemistry and Electrochemistry, Russian Academy of Sciences, Moscow, Russia
- Daniel Goldberg, MD, PhD, Washington University St. Louis, St. Louis, MO
- Samuel T. Hess, PhD, University of Maine, Orono, ME
- Sukbir Kaur, PhD, Laboratory of Pathology, Center for Cancer Research, NCI, Bethesda, MD
- Leonid Margolis, PhD, Section on Intercellular Interactions, NICHD, Bethesda, MD
- Doreen Matthies, PhD, Unit on Structural Biology, NICHD, Bethesda, MD
- Ana Olivera, PhD, Mast Cell Biology Section, NIAID, Bethesda, MD
- Richard Pastor, PhD, Laboratory of Membrane Biophysics, NHLBI, Bethesda, MD
- Annika Pfeiffer Daniels, PhD, Laboratory of Allergic Diseases, NIAID, Bethesda, MD
- Thomas S. Reese, MD, Laboratory of Neurobiology, NINDS, Bethesda, MD
- David D. Roberts, PhD, Laboratory of Pathology, Center for Cancer Research, NCI, Bethesda, MD
- Santimukul Santra, PhD, Pittsburg State University, Pittsburg, KS
- Anna Shnyrova, PhD, Universidad del País Vasco, Bilbao, Spain
- Alexander J. Sodt, PhD, Unit on Membrane Chemical Physics, NICHD, Bethesda, MD
- Niraj H. Tolia, MD, PhD, Laboratory of Malaria Immunology and Vaccinology, NIAID, Bethesda, MD
- Gary R. Whittaker, PhD, College of Veterinary Medicine, Cornell University, Ithaca, NY
- Jack Yanovski, MD, PhD, Section on Growth and Obesity, NICHD, Bethesda, MD
- Wei Zheng, PhD, Division of Preclinical Innovation, NCATS, Bethesda, MD
- Fei Zhou, PhD, Unit on Structural Biology, NICHD, Bethesda, MD
Contact
For more information, email zimmerbj@mail.nih.gov or visit https://irp.nih.gov/pi/joshua-zimmerberg.