Biophysics of Large Membrane Channels

- Sergey M. Bezrukov, PhD, DSci, Head, Section on Molecular Transport
- Tatiana K. Rostovtseva, PhD, Associate Scientist
- Philip A. Gurnev, PhD, Staff Scientist
- María Queralt-Martín, PhD, Visiting Fellow
- Megha Rajendran, PhD, Visiting Fellow
- William M. Rosencrans, BS, Intramural Research Training Award Fellow
We study mitochondrial and bacterial membrane proteins that form “large” beta-barrel channels responsible for metabolite fluxes between cells and cellular compartments. Healthy cell functioning and development require effective communication between cells and cell organelles, which is facilitated by beta-barrel membrane channels. We are interested in the physical mechanisms that regulate such channels under normal and pathological conditions. Among many wet-lab approaches, such as fluorescence correlation spectroscopy, bilayer overtone analysis, and confocal microscopy, our hallmark method is to reconstitute channel-forming proteins into planar lipid membranes, which allows us to study them at the single-molecule level. Empirical findings obtained in these experiments are rationalized within a framework of a physical theory of channel-facilitated transport, which brings an understanding necessary to design new strategies to effectively correct the deviant interactions associated with disease.
In contrast to the highly ion-selective channels studied in neurophysiology, which have narrow selectivity filters to match the size of partially dehydrated ions, metabolite, or “nutrient,” channels are significantly wider. Indeed, they have to accommodate metabolite molecules that are typically much larger than simple mono- or divalent ions. Because of their size and their primarily beta-barrel scaffolds, the mechanisms of the selectivity and gating of such channels are quite different from those of the ion-selective channels formed from alpha-helical subunits. To grasp the general principles of beta-barrel channel functioning and regulation, we work with a variety of proteins and channel-forming peptides.
The channel-forming proteins and peptides we work with include VDAC (voltage-dependent anion channel from the outer membrane of mitochondria), alpha-hemolysin (toxin from Staphylococcus aureus), translocation pores of Bacillus anthracis (PA63), Clostridium botulinum (C2IIa), and C. perfringens (Ib) binary toxins, Epsilon toxin (from C. perfringens), OmpF (general bacterial porin from Escherichia coli), LamB (sugar-specific bacterial porin from E. coli), OprF (porin from Pseudomonas aeruginosa), MspA (major outer-membrane porin from Mycobacterium smegmatis), Alamethicin (amphiphilic peptide toxin from Trichoderma viride), Syringomycin E (lipopeptide toxin from Pseudomonas syringae), and the bacterial peptide TisB involved in persister cell formation. We also use gramicidin A (linear pentadecapeptide from B. brevis) as a molecular sensor of membrane mechanical properties. With the goal of studying the channel-forming proteins under controlled conditions, we first isolate them from the host organisms, purify them, and then reconstitute them into planar lipid bilayers, the model system with precisely defined physical properties. This allows us to explore channel interactions with the lipid membrane as modified by volatile anesthetics, cytosolic proteins, such as tubulin and alpha-synuclein, and newly synthesized drugs that act as blockers of the translocation pores of bacterial toxins. Our motivation is that learning the molecular mechanisms of channel functioning is vital for developing new approaches for the treatment of various diseases, for which regulation of transport through ion channels plays the key role.
Click image to view.
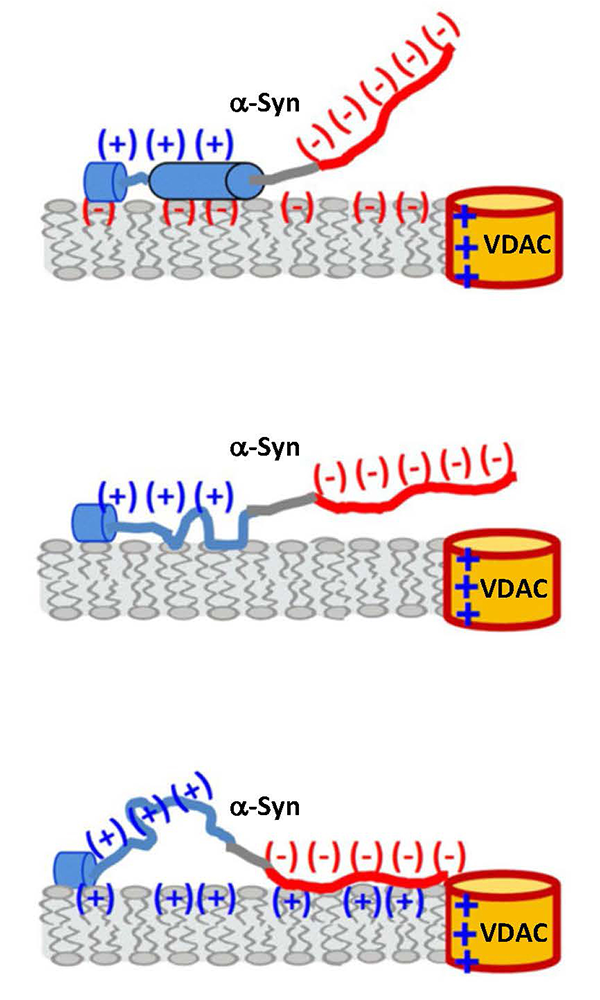
Figure 1. Membrane-catalyzed alpha-synuclein interaction with VDAC
A tentative model qualitatively accounting for the electrostatic and hydrophobic interactions and conformational variability of α-syn on the membrane surface. The availability of the anionic C-terminal domain of α-syn for VDAC pore blockage depends on the membrane-lipid charge and salt concentration. Interaction of each α-syn domain with the membrane affects the molecule’s overall membrane affinity, as well as the rate of capture of the C-terminal domain by the VDAC nanopore. The membrane-binding region is shown in blue (net 3 positive charges), nonpolar NAC (non-amyloid-β component) domain in grey, and the anionic C-terminal domain in red (net 15 negative charges). Anionic lipids enhance α-syn binding and promote the formation of two helical membrane-binding domains in the N-terminus, but they electrostatically repel the anionic C-terminal domain from the membrane surface, reducing its availability for the capture by the nanopore. For zwitterionic, and perhaps cationic lipids, a more random configuration of the N-terminus can be expected, resulting in multiple α-syn conformations on the membrane surface.
Membrane association of alpha-synuclein domains studied using VDAC nanopore reveals an unexpected binding pattern.
The exceptional diversity of interactions between peripheral membrane proteins and bilayer lipid membranes makes the membrane surface a rich scene for cellular functions and their regulation. However, this diversity presents significant experimental barriers to the studies of interaction mechanisms among the components. One of the main challenges is that the energies of interaction between individual protein residues and lipid molecules are small; thus, statistical effects play a significant role. A variety of techniques to characterize binding of peripheral membrane proteins to liposome or planar lipid bilayer platforms have been developed, but it is increasingly clear that, except in the simplest of systems, no isolated technique yields a clear picture of the membrane binding process. In part, this is because these assays measure the average binding parameters of an ensemble of molecules, highlighting the need to characterize membrane-bound proteins at the single-molecule level.
This year, we used the voltage-dependent anion channel (VDAC) of the outer mitochondrial membrane as a single-molecule probe for alpha-synuclein (α-syn), a protein of considerable clinical interest owing to its well-established involvement in the pathology of Parkinson's disease. It is well-established that α-syn binding from solution to the surface of membranes composed of negatively charged and/or non-lamellar lipids can be characterized by equilibrium dissociation constants in the order of tens of micromolar. However, we previously found that VDAC, reconstituted into planar bilayers of a plant-derived lipid, responds to α-syn at nanomolar solution concentrations. Now, using lipid mixtures that mimic the composition of mitochondrial outer membranes, we showed that functionally important binding does indeed take place in the nanomolar range. We demonstrated that the voltage-dependent rate at which a membrane-embedded VDAC nanopore captures α-syn is a strong function of membrane composition. Comparison of the nanopore results with those obtained by the bilayer overtone analysis of membrane binding demonstrated a pronounced correlation between the two datasets. The stronger the binding, the larger the on-rate, but with some notable exceptions. This leads to a tentative model of α-syn–membrane interactions, which assigns different lipid-dependent roles to the N- and C-terminal domains of α-syn, accounting for both electrostatic and hydrophobic effects. As a result, the rate of α-syn capture by the nanopore is not simply proportional to the α-syn concentration on the membrane surface but sensitive to the specific interactions of each domain with the membrane and nanopore.
Click image to view.
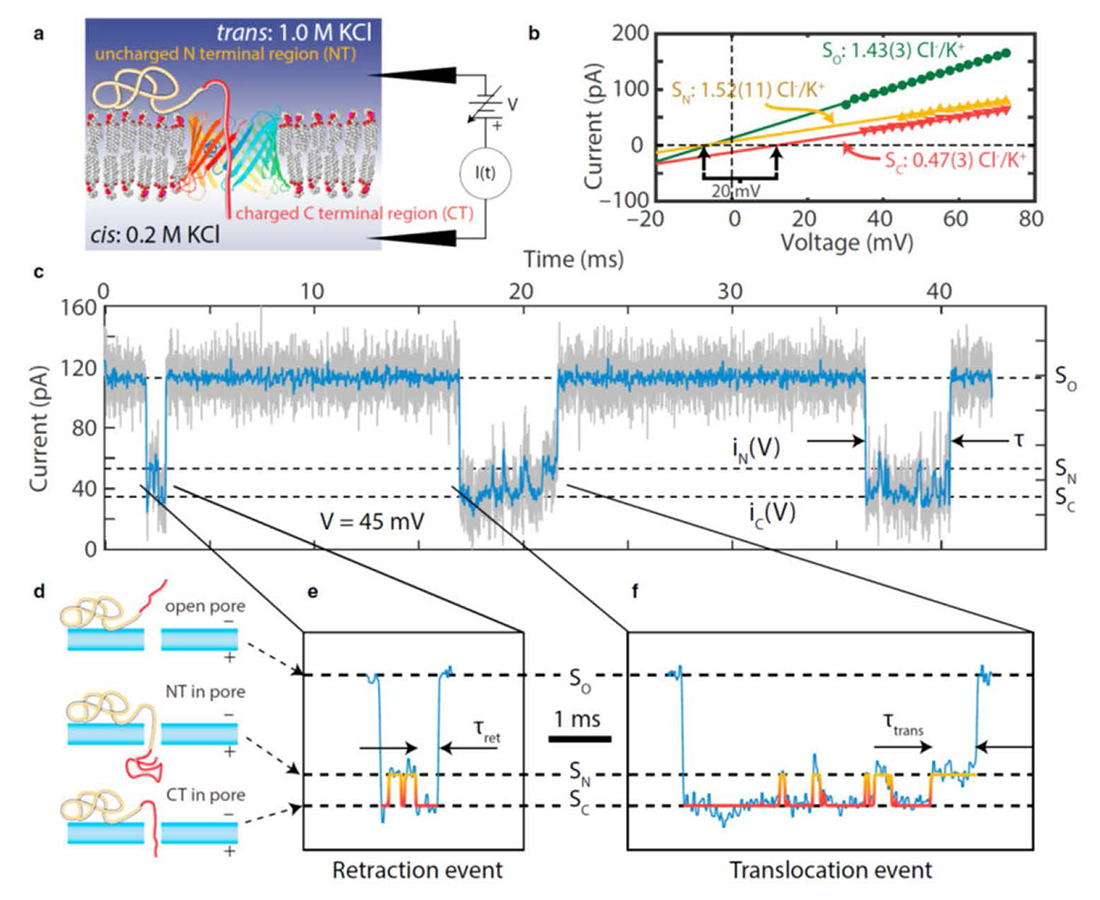
Figure 2. Watching translocation of single proteins
Salt concentration gradient–enhanced observation of α-syn dynamics in a VDAC nanopore.
a. Experimental setup (not to scale). The “diblock-copolymer”–like structure of α-syn consists of two “selectivity tags” of differing charge density (light yellow and dark red) that modulate the electrical properties of the VDAC nanopore differently under a salt concentration gradient.
b. Current-voltage curves of the open pore high-conducting (SO, dark green circles) and the two low-conducting substates when α-syn is inside the pore (SN, yellow triangles and SC, red inverted triangles); selectivities were calculated from the reversal potentials (vertical arrows).
c. Sample current record; as-recorded data are shown in light gray and software-filtered data in dark blue.
d. Identification of the substates by noting that the capture of the C-terminus in the nanopore corresponds to the lower-conducting substate.
e. and f. Details of a retraction (e) and a translocation (f) event.
Real-time nanopore-based recognition of protein translocation success
A growing number of new technologies are supported by a single- or multi-nanopore architecture for capture, sensing, and delivery of polymeric biomolecules. Nanopore-based single-molecule DNA sequencing is the premier example. The method relies on the uniform linear charge density of DNA, so that each DNA strand is overwhelmingly likely to pass through the nanopore and across the separating membrane. By contrast, translocation is not assured for disordered peptides, folded proteins, or block copolymers with heterogeneous charge densities, and additional strategies to monitor the progress of the polymer molecule through a nanopore are required. This year, we studied the translocation of a heterogeneously charged polypeptide using model-free detection of the effect of the polymer charge on the electrical environment inside the nanopore to monitor the translocation process of single polypeptides in real time. This was accomplished using “selectivity tags,” regions of different but uniform charge density at the ends of a polypeptide, that produce different selectivity of the nanopore to cations and anions, and hence ionic current levels, in a voltage-biased nanopore under a salt concentration gradient. We employed the natural, disordered “diblock copolymer”–like 140 amino-acid polypeptide α-syn, which comprises two such tags, a highly negatively charged C-terminal region (CT; 43 amino acids, total charge –15e) and a largely neutral N-terminal region (NT; 97 amino acids, total charge +3e). By using these features, we demonstrated a single-molecule method for direct, model-free, real-time monitoring of the translocation of a disordered, heterogeneously charged polypeptide through a nanopore. The two selectivity tags at the ends of the polypeptide enabled us to discriminate between α-syn translocation and retraction. Our results demonstrated exquisite sensitivity of polypeptide translocation to the applied transmembrane potential and proved the principle that nanopore selectivity reports on biopolymer substructure. We anticipate that the selectivity tag technique will be broadly applicable to nanopore-based protein detection, analysis, and separation technologies, and to the elucidation of protein translocation processes in normal cellular function and in disease.
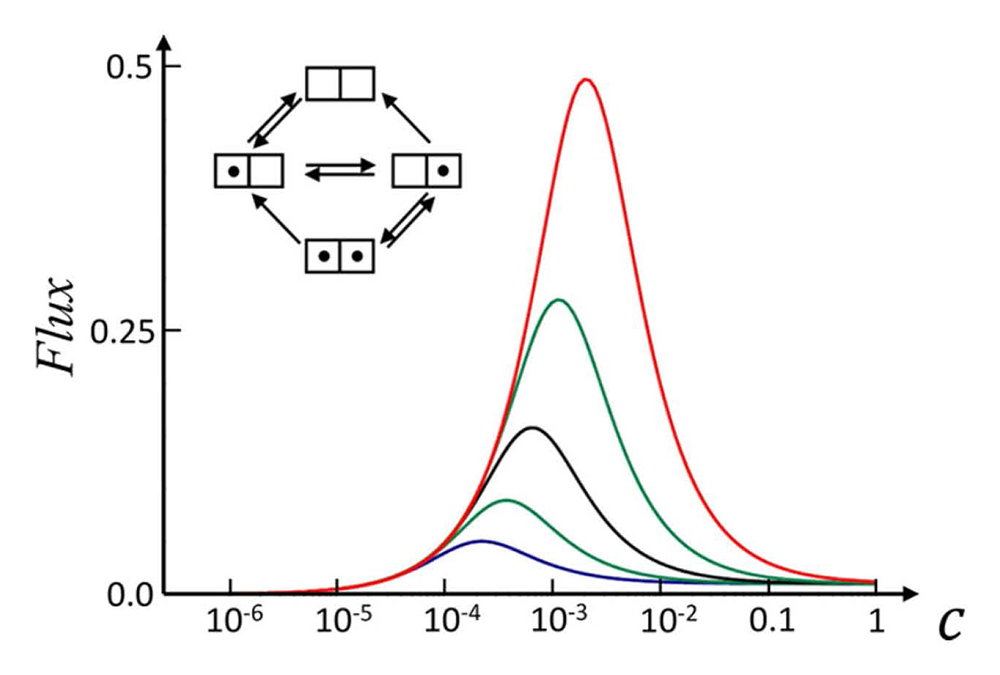
Mapping intra-channel diffusive dynamics of interacting molecules onto a two-site model: crossover in flux concentration dependence
Transport of various solutes through membrane channels is an extremely complex phenomenon. One of the reasons for this complexity is interactions of solute molecules between themselves and with the channel. This year, we addressed this problem by analyzing how such interactions affect the flux dependence on the solute concentration. The study focused on narrow membrane channels, for which it was assumed that the molecules cannot bypass each other because of their hard-core repulsion. In addition, other short- and long-range solute-solute interactions were also considered. Such interactions make it impossible to develop an analytical theory for the flux in the framework of the continuum diffusion model of solute dynamics in the channel, developed in our lab during recent years. To overcome this difficulty, we coarse-grained the diffusion model by mapping it onto a two-site one where the rate constants describing the solute dynamics were expressed in terms of the parameters of the initial diffusion model. This allowed us, first, to find an analytical solution for the flux as a function of the solute concentration and, second, to characterize the solute-solute interactions by two dimensionless parameters. Such a characterization proved to be very informative, as it resulted in a clear classification of the effects of the solute-solute interactions on the concentration dependence of the flux. Unexpectedly, it turned out that this dependence can be nonmonotonic, exhibiting a sharp maximum as a function of system parameters. In other words, we found that the effect is quite nontrivial: the flux can reach a well-pronounced maximum at a certain “optimal” concentration, the value of which is defined by the interaction parameters. We hypothesize that this phenomenon may be used by nature as a sensory mechanism of a regulatory circuit, wherein an optimal solute concentration is reported upon by maximizing the transmembrane flux of the molecules.
Click image to view.
Figure 3. Crossover in flux concentration dependence
A simple two-site model of channel-facilitated membrane transport allows for a description of not only flux saturation with the solute concentration (C) but also more subtle effects, such as superlinearity and crossover behavior. In particular, solute attraction to the channel walls in combination with strong short-range solute-solute attraction may result in a sharp peak in the concentration dependence of the flux. Different curves correspond to different strengths of solute-solute interactions, increasing from bottom to top. This rich behavior may play a role in both sensory and regulation mechanisms in cellular metabolic circuits.
Additional Funding
- Colgate University award to William Rosencrans
Publications
- Hoogerheide DP, Gurnev PA, Rostovtseva TK, Bezrukov SM. Real-time nanopore-based recognition of protein translocation success. Biophys J 2018;114:772–776.
- Berezhkovskii AM, Bezrukov SM. Mapping intra-channel diffusive dynamics of interacting molecules onto a two-site model: crossover in flux concentration dependence. J Phys Chem B 2018;122:10996-11001.
- Jacobs D, Hoogerheide DP, Rovini A, Jiang Z, Lee JC, Rostovtseva TK, Bezrukov SM. Probing membrane association of alpha-synuclein domains with VDAC nanopore reveals unexpected binding pattern. Sci Reports 2019;9:4580.
- Queralt-Martín M, Bergdoll L, Jacobs D, Bezrukov SM, Abramson J, Rostovtseva TK. Assessing the role of residue E73 and lipid headgroup charge in VDAC1 voltage gating. Biochim Biophys Acta – Bioenerg 2019;1860:22–29.
- Berezhkovskii AM, Dagdug L, Bezrukov SM. Exact solutions for distributions of first-passage, direct-transit, and looping times in symmetric cusp potential barriers and wells. J Phys Chem B 2019;123:3786–3796.
Collaborators
- Vicente M. Aguilella, PhD, Universidad Jaume I, Castellón, Spain
- Alexander M. Berezhkovskii, PhD, Division of Computational Bioscience, CIT, NIH, Bethesda, MD
- Susan K. Buchanan, PhD, Laboratory of Molecular Biology, NIDDK, Bethesda, MD
- Leonid V. Chernomordik, PhD, Section on Membrane Biology, NICHD, Bethesda, MD
- Leonardo Dagdug, PhD, Universidad Autónoma Metropolitana-Iztapalapa, Mexico City, Mexico
- David Hoogerheide, PhD, National Institute of Standards and Technology, Gaithersburg, MD
- Jennifer C. Lee, PhD, Biochemistry and Biophysics Center, NHLBI, Bethesda
- Ekaterina M. Nestorovich, PhD, The Catholic University of America, Washington, DC
- Sergei Y. Noskov, PhD, University of Calgary, Calgary, Canada
- Olga Protchenko, PhD, Liver Diseases Branch, NIDDK, Bethesda, MD
- Dan Sackett, PhD, Section on Cell Biophysics, NICHD, Bethesda, MD
- Alexander Sodt, PhD, Unit on Membrane Chemical Physics, NICHD, Bethesda, MD
- Gerhard Wagner, PhD, Harvard Medical School, Cambridge, MA
- Michael Weinrich, MD, Office of the Director, NICHD, Bethesda, MD
- David L. Worcester, PhD, National Institute of Standards and Technology, Gaithersburg, MD
- Joshua Zimmerberg, MD, PhD, Section on Integrative Biophysics, NICHD, Bethesda, MD
Contact
For more information, email bezrukov@helix.nih.gov or visit http://smt.nichd.nih.gov.


