Neuronal Circuits Controlling Behavior: Genetic Analysis in Zebrafish

- Harold Burgess, PhD, Head, Section on Behavioral Neurogenetics
- Tripti Gupta, PhD, Staff Scientist
- Ashwin A. Bhandiwad, PhD, Postdoctoral Fellow
- Eric J. Horstick, PhD, Postdoctoral Fellow
- Jennifer Panlilio, PhD, Postdoctoral Fellow
- Abhignya Subedi, PhD, Postdoctoral Fellow
- Svetlana Semenova, PhD, Visiting Fellow
- Jennifer L. Sinclair, MSc, Zebrafish Technician
- Nickolas Chu, BSc, Postbaccalaureate Fellow
- Jacob Clarin, BSc, Postbaccalaureate Fellow
- Victoria Heigh, BSc, Postbaccalaureate Fellow
Our goal is to understand how, under diverse environmental contexts, the nervous system selects appropriate behavioral responses to sensory information in a way that best satisfies internal motivational objectives. We use the larval zebrafish as a model because its brain exhibits the basic architecture of the vertebrate brain but is much less complex than the mammalian brain. Despite the relative simplicity of their nervous system, zebrafish have a sophisticated repertoire of sensory-guided and internally driven behaviors. Furthermore, the optical clarity of the embryo facilitates visualization of individual neurons and their manipulation with genetic techniques. Behavior in larvae is innate and therefore exhibits minimal variability between fish. Subtle alterations in behavior can therefore be robustly measured, making it possible to quickly assess the contribution of identified neurons to a variety of motor behaviors.
We focus on two aspects of behavioral regulation: the neuronal mechanisms by which sensory context regulates behavioral decisions and the pathways that sustain changes in behavioral state. Neuronal connections that allow the brain to integrate sensory and internal-state information are established through genetic interactions during development, and are frequently disrupted by gene mutations associated with neurodevelopmental disorders. We are therefore able to use discoveries about sensorimotor integration pathways to understand how human disease genes disrupt brain development. Finally, to support these objectives, we develop new genetic tools and behavioral assays to probe the nexus between neuronal function and behavior at single-cell resolution.
Neuronal pathways for auditory sensory processing
Startle responses are rapid reflexes that are triggered by sudden sensory stimuli and which help animals defend against or escape from potentially threatening stimuli. In both fish and mammals, startle responses are initiated by giant reticulospinal neurons in the medulla, which receive short-latency sensory input from diverse sensory modalities. Although highly stereotyped, startle responses are nevertheless modulated by sensory context and behavioral state and are therefore an excellent system in which to understand how such information is integrated for behavioral choice. In mammals, including humans, the startle response to a strong auditory stimulus can be inhibited by pre-exposure to a weak acoustic ‘prepulse.’ This form of startle modulation, termed prepulse inhibition (PPI), is diminished in several neurological conditions. We aim to resolve the core neuronal pathway that mediates PPI to then understand how gene mutations linked to neurodevelopmental disorders disrupt sensory processing.
Vibrational stimuli trigger rapid-escape swims in zebrafish, which are mediated by giant reticulospinal neurons, in a manner similar to the central neurons controlling startle responses in mammals. Escape swims are suppressed by preexposure to a prepulse, allowing us to apply the powerful suite of genetic tools available in zebrafish to identify neurons that mediate prepulse inhibition. To identify a transgenic zebrafish line that genetically labels neurons required for PPI, we screened a library of neuron-specific Gal4–enhancer trap lines marking distinct populations of neurons in the brain, by ablating the neurons in each enhancer trap line before testing for PPI. The transgenic line y252 labels a discrete population of neurons in the hindbrain whose ablation or optogenetic inhibition eliminates PPI. The neurons labeled in y252 are specified by the transcription factor gsx1. To identify the precise subset of gsx1 neurons that mediate PPI, we then used volumetric calcium imaging to simultaneously visualize the activity in tens of thousands of gsx1 neurons during PPI. The method enabled us to locate a specific brain region that contains gsx1 neurons, whose activity correlates strongly with behavioral PPI. To determine where such neurons are causally related to PPI, we optogenetically activated them before probing fish with a startle stimulus. Controlled firing of gsx1 neurons was able to reproduce the effect of a prepulse.
We next visualized the morphology and projections of such neurons in order to reveal how they connect to the core startle circuit. For this, we developed a new method that exploits the low efficiency of the B3 recombinase in zebrafish to achieve stochastic labeling of isolated neurons in cells that co-express the Gal4 transcription factor and the Cre recombinase. We found a specific subset of gsx1 neurons that use the neurotransmitter glutamate and that project to an area of the lateral dendrite of the Mauthner cell, the command neuron for startle responses in fish, which receives auditory input. By directly visualizing neurotransmitter release from the auditory nerve, we discovered that acoustic information to the Mauthner cell is selectively depressed during PPI, while transmission to other brain regions is spared, indicating that a key mechanism for PPI is presynaptic inhibition [Reference 1]. Given that PPI is abnormal in neuropsychiatric disorders with developmental origins, including schizophrenia and autism, our work will help identify and probe fundamental defects in circuitry that are abnormal in these conditions.
As in humans, zebrafish have more than one pathway for escape behavior. At larval stages, auditory stimuli drive either an extremely rapid Mauthner cell–mediated response, or a slightly delayed non-Mauthner behavior. Delayed responses are of special interest as they integrate additional information, including visual cues, into decisions on escape trajectory, but almost nothing is known about the neuronal pathway that mediates such behavior. We performed a circuit-breaking screen to identify neurons that initiate delayed escape and discovered a population of cells adjacent to the cerebellum in the prepontine hindbrain that initiate delayed escapes. Auditory cues that are insufficiently threatening to drive an immediate escape behavior activate the prepontine delayed escape population, allowing larvae to perform a flexible and graded response (Figure 1). The identification of such neurons provides a new paradigm for dissecting the circuit that mediates very rapid behavioral selection in an ethologically critical pathway [Reference 2].
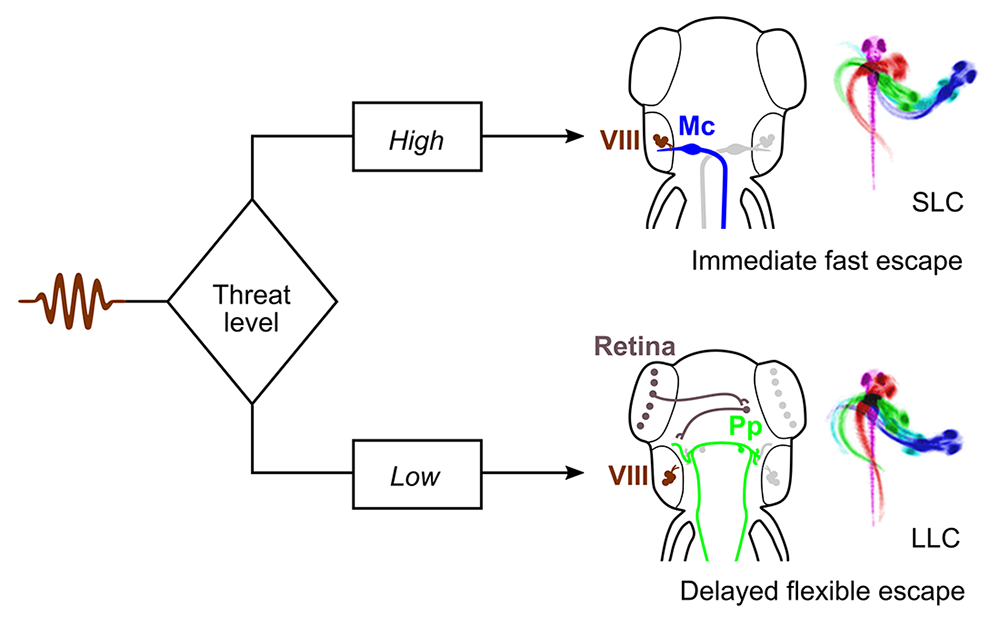
Click image to view.
Figure 1. Behavioral selection in the auditory escape pathway
Threat levels select between rapid Mauthner cell (Mc)–driven escape responses and delayed flexible escape behavior initiated by a cluster of neurons in the prepontine hindbrain (Pp).
Tools for decoding neuronal circuits
During our work to identify the neuronal basis for prepulse inhibition, we realized that existing transgenic tools did not permit manipulation of small groups of neurons with a high degree of specificity. We developed a system to leverage the thousands of Gal4 lines already available in zebrafish, by generating a new library of Cre enhancer trap lines with restricted patterns of expression within the brain. The lines can be used 'intersectionally' to restrict the pattern of expression with Gal4 domains, allowing small clusters of neurons to be targeted. Thus, during our studies on prepulse inhibition, we used a subset of the lines to ablate gsx1 neurons within specific hindbrain domains to determine which specific neurons are required for prepulse inhibition. A unique feature of brain imaging in zebrafish is the ability to visualize the total architecture of the brain while simultaneously recording the position of every constituent labeled neuron. Brain registration techniques enable data from several individual zebrafish to be co-aligned to very high precision. Thus after generating our new toolkit of Cre lines, we performed whole-brain imaging for each line, then registered the image of each line to the same reference brain. We then developed a new online brain atlas for larval zebrafish that enables researchers to quickly visualize the larval brain and locate transgenic lines to aid experiments (Figure 2). The Zebrafish Brain Browser leverages the power of Web3D to provide 3D visualization of the zebrafish brain within any web browser [Reference 3].
We postulated that the very high precision of alignment may enable statistically robust whole-brain analysis of neuronal composition and morphology in zebrafish models of neurological disorders. For this, we first developed a new computational method to automatically segment the larval zebrafish brain into 180 neuroanatomical regions, based on gene expression patterns. We then generated new software, Cobraz, that uses those regions to identify changes in brain size, neurotransmitter distribution, or local morphology in confocal images of genetic mutant zebrafish larvae. We validated our algorithm by comparing embryos injected with the atoh7 morpholino to wild-type siblings. Loss of atoh7 is known to eliminate retinofugal projections. Indeed, we found that the optic tectum, the major retinorecipient area, was significantly smaller in morphants [Reference 4]. The technique can be applied to almost any zebrafish neurodevelopmental model, thus enabling robust and quantitative detection of subtle changes in brain structure or composition.
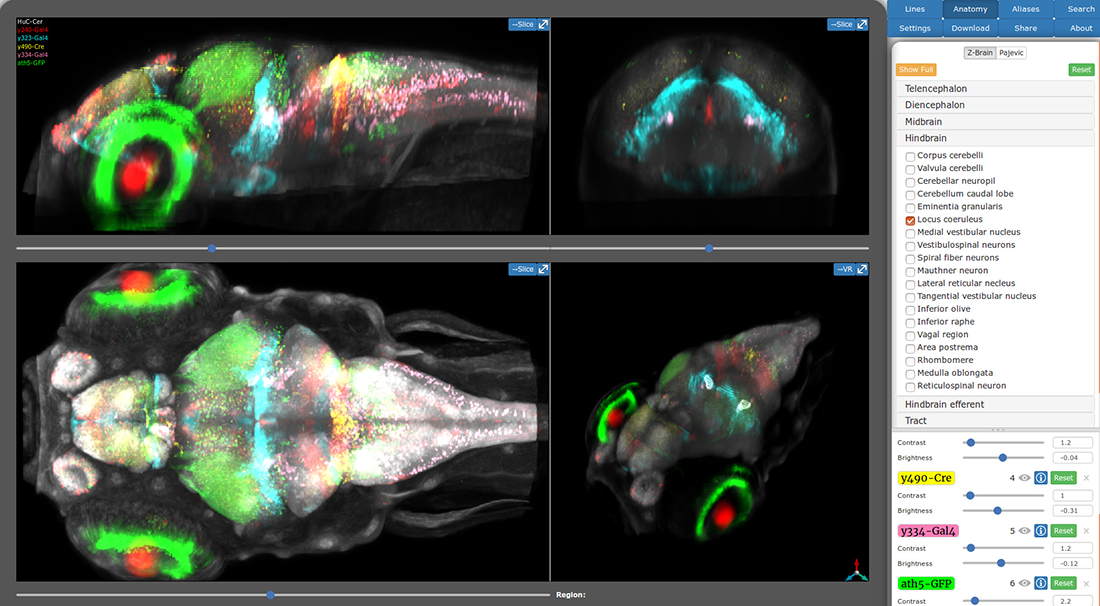
Click image to view.
Figure 2. Zebrafish Brain Browser Web atlas
The new online Zebrafish Brain Browser visualizes transgene expression in three dimensions: by enabling users to toggle between projections through selected brain regions, or to view single slices through the brain in each cardinal axis. The atlas comprises almost 300 expression patterns, facilitating selection of transgenic lines to experimentally manipulate selected neurons within the larval brain.
Additional Funding
- DIR Director's Award, NICHD (2018-2019) with Dr. David Clark, NICHD
- DIR Director's Award, NICHD (2018-2019) with Dr. Karl Pfeifer, NICHD
- Simons Foundation Autism Research Initiative
Publications
- Tabor KM, Smith TS, Brown M, Bergeron SA, Briggman KL, Burgess HA. Presynaptic inhibition selectively gates auditory transmission to the brainstem startle circuit. Curr Biol 2018;28(16):2527-2535.
- Marquart GD, Tabor KM, Bergeron SA, Briggman KL, Burgess HA. Prepontine non-giant neurons drive flexible escape behavior in zebrafish. PLoS Biol 2019;17:e3000480.
- Tabor KM, Marquart GD, Hurt C, Smith TS, Geoca AK, Bhandiwad AA, Subedi A, Sinclair JL, Rose HM, Polys NF, Burgess HA. Brain-wide cellular resolution imaging of Cre transgenic zebrafish lines for functional circuit-mapping. eLife 2019;8:e42687.
- Gupta T, Marquart GD, Horstick EJ, Tabor KM, Pajevic S, Burgess HA. Morphometric analysis and neuroanatomical mapping of the zebrafish brain. Methods 2018;150:49-62.
- Shainer I, Michel M, Marquart GD, Bhandiwad AA, Zmora N, Livne ZBM, Zohar Y, Hazak A, Mazon Y, Förster D, Hollander-Cohen L, Cone RD, Burgess HA, Gothilf Y. Agouti-related protein 2 is a new player in the teleost stress response system. Curr Biol 2019;29:2009-2019.
Collaborators
- Kevin Briggman, PhD, Research Center caesar (center of advanced european studies and research), Max Planck Institute for Metabolism Research, Bonn, Germany
- Yoav Gothilf, PhD, Tel-Aviv University, Tel-Aviv, Israel
- Sinisa Pajevic, PhD, Mathematical and Statistical Computing Lab, CIT, NIH, Bethesda, MD
- Nicholas Polys, PhD, Advanced Research Computing, Department of Computer Science, Virginia Polytechnic Institute and State University, Blacksburg, VA
Contact
For more information, email haroldburgess@mail.nih.gov or visit http://ubn.nichd.nih.gov.


