Gene Regulation in Innate Immunity

- Keiko Ozato, PhD, Head, Section on Molecular Genetics of Immunity
- Anup Dey, PhD, Biologist
- Tiyun Wu, PhD, Staff Scientist
- Sakshi Chauhan, PhD, Visiting Fellow
- Lili Chen, PhD, Visiting Fellow
- Eri Hosogane, PhD, Visiting Fellow
- Vishal Nehru, PhD, Visiting Fellow
- Keita Saeki, MD, PhD, Visiting Fellow
- Richard Pan, BS, Postbaccalaureate Fellow
Macrophages and related cells such as microglia recognize incoming pathogens and produce cytokines, notably interferons (IFNs), the interleukins IL-1 and IL-6, and tumor necrosis factor alpha (TNF-alpha). While IFNs impart antiviral and antimicrobial protection to the host, the latter cytokines are associated with inflammatory responses. IFNs are produced upon activation of the IRF (interferon regulatory factor) family of transcription factors, while inflammatory cytokines are produced by activation of the transcription factor NFκB. Our goal is to study the molecular pathways that direct the development and function of macrophages and other myeloid cells. To this end, we focus on the role of IRF8 in innate immunity. IRF8, a member of the IRF family, is expressed at high levels in macrophages, microglia, and dendritic cells (DCs), and is required for the production of both type I and type II IFNs. IRF8 is essential for mounting the first line of defense against various invading pathogens prior to the initiation of antigen-specific adaptive immune responses.
Transcriptionally active genes are embedded in chromatin that is dynamically exchanged, whereas silenced genes are surrounded by more stable chromatin. The chromatin environment contributes to the epigenetic states of given cells and influences transcriptional processes. We have long been working on BRD4, a bromodomain protein that binds to acetylated histones and promotes active transcription. BRD4 is involved in the dynamic chromatin exchange that takes place in highly transcribed genes, an exchange that requires a special histone called H3.3. As a result of the association with transcription, H3.3 is implicated in epigenetic control of gene expression patterns. Our goal is to elucidate the activity of BRD4 and histone H3.3 in innate immunity.
The role of IRF8 in brain inflammation: Alzheimer's disease and Aicardi-Goutières syndrome
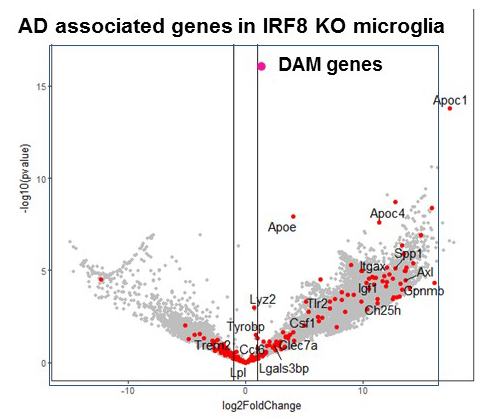
IRF8 is a transcription factor of the conserved interferon regulatory factor family. It is expressed in cells important for host antipathogen protection and is involved in inflammatory responses. Recent evidence shows that, through its expression in microglia, IRF8 plays a significant role in inflammation in the brain. SNP (single nucleotide polymorphism)–based genome-wide association studies revealed that inflammatory genes expressed in microglia are risk factors for Alzheimer's disease (AD), which causes memory loss followed by breakdown in broader cognitive function. We have begun to study the role of IRF8 in AD using the mouse model of AD. Histological studies found that microglia scattered over the entire brain have abnormal morphology in IRF8 knockout (KO) mice (Figure 1). IRF8 KO microglia were all devoid of extensive dendrites projecting next to neurons and appeared similar to those in AD models. RNA-seq analysis of wild-type (WT) and IRF8 KO microglia showed that a large array of AD–associated genes were activated in IRF8 KO microglia (Figure 2). Some of the genes (red dots) were found to be AD risk factors, including ApoE (encoding apolipoprotein E), which represented the highest risk factor.
Click image to view.
Figure 1. Alzheimer's disease–associated genes in IRF8 KO microglia
Microglia were sorted from wild-type and IRF8 KO brain (3-month-old), and RNA-seq (an RNA sequencing method) analyses were performed. Genes upregulated in IRF8 KO microglia were enriched with genes upregulated in microglia of the Alzheimer's disease mouse model.
Aicardi-Goutières syndrome (AGS) patients present with various forms of encephalopathy with prominent neuroinflammation. Symptoms include vision/motor deficits, cognitive deficiency, and vascular damage in the brain associated with lupus (SLE)–like autoimmune conditions. Classically, AGS has been linked to mutations in enzymes that degrade endogenous nucleic acids (DNA and RNA), produced by normal biological processes, including replication, transcription, and DNA repair [Crow YJ, Manel N. Nat Rev Immunol 2015;15:429]. Mutations in any of the three RNase H2 subunits account for those in more than 50% of AGS patients. Mutations in other nucleic acid–metabolizing factors linked to AGS are in TREX1 (encoding three-prime repair exonuclease 1), ADAR (encoding double-stranded RNA–specific adenosine deaminase), and SAMHD1 (encoding deoxynucleoside triphosphate triphosphohydrolase). More recently, constitutively activating mutations in the signaling pathways that sense excess nucleic acids and activate innate immune responses have been found to also cause AGS–like diseases. The Crouch lab showed that defects in RNase H2 lead to accumulation of DNA and activate the cGAS-STING pathway, which detects the presence of cytosolic DNA; loss of Sting eliminates activation of interferon-stimulated genes (ISGs). Accumulation of RNA triggers the RIG-I/MDA5 pathway, which plays a major role in sensing RNA–virus infection by detecting cytoplasmic viral double-stranded RNA. Both pathways signal Tbk1 (a serine/threonine kinase that plays an essential role in regulating inflammatory responses to foreign agents), leading to activation of type I interferons and their downstream pathways. Thus, activation of ISGs, coined the "interferon signature," is a defining feature of AGS. ISG proteins are found in spinal fluids, presumably produced in microglia (although not proven), and in peripheral monocytes/macrophages. Recent developments in the field have broadened the description of AGS with regard to its onset, phenotypic severity, and variability. Therefore, in collaboration with Robert Crouch and Yoh-suke Mukouyama, we are studying how IRF8 affects microglial inflammatory responses in mice carrying the mutation in the RNAseH2A, which causes AGS in humans.
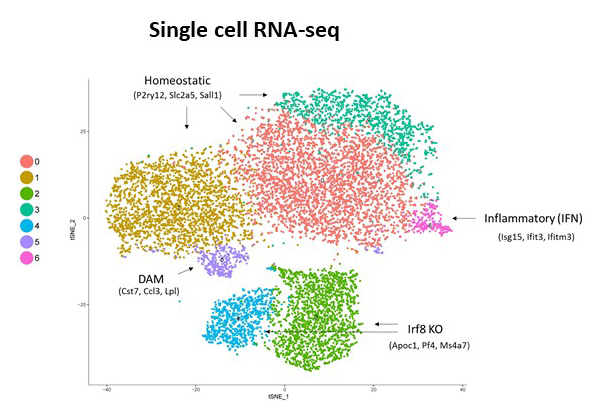
To further investigate the role of IRF8 in AD, we crossed IRF8KO mice with 5xFAD mice, a model of AD. We found that amyloidβ accumulation was reduced in 5xFAD microglia when crossed with IRF8KO. Consistent with these data, we found that the lysosomal activation marker CD68 was less in the absence of IRF8 (Figure 1). With the help of Steven Coon, we performed single cell RNA-seq for microglia from these mice. Our results indicated that IRF8KO microglia form a cell population with transcriptome profiles distinct from those of WT or 5xFAD. Our analysis indicated that IRF8KO and AD mice share transcriptome profiles more closely than WT (Figure 2).
Click image to view.
Figure 2. Single-cell RNA-seq was performed with the 10X genomics system.
Seven clusters were identified among WT, IRF8 KO, 5xFAD, and the cross of the latter two. IRF8KO microglia show a unique cluster. DAM: disease-associated microglia genes
Context-dependent role for BRD4
RD4 is a bromodomain protein of the BET (bromodomain and extraterminal domain) family, which this laboratory has been studying for many years. BRD4 is expressed at high levels in most, if not all cells, and is necessary for very early embryonic development. Thus, conventionally created Brd4 knockout (KO) mice are embryonic lethal. BRD4 is a so-called “chromatin reader” owing to its binding to acetylated histones. It also recruits the transcription elongation factor P-TEFb, thus facilitating transcriptional elongation. Moreover, BRD4 plays a critical role in forming superenhancers. Superenhancers are long stretches of regulatory DNAs densely occupied by transcription factors and chromatin regulators. They direct strong transcription of select genes and thus help define cellular and lineage identity. In the past several years, research on BRD4 has seen a dramatic upturn owing to the development of small-molecule inhibitors that inhibit binding of acetyl-histones to the BET family proteins. These inhibitors, affecting mostly the BET protein BRD4, antagonize cancer growth, particularly leukemia and lymphoma. Furthermore, BET inhibitors have been shown to inhibit inflammatory responses related to cardiovascular and autoimmune diseases. Such reports implicate BRD4 in various disease processes and offer new therapeutic possibilities for several difficult-to-treat illnesses; indeed, clinical trials are being conducted for leukemia and inflammation. However, the developments present new issues stemming from the dearth of our understanding of the precise role of BRD4 in health and disease and of the mechanism of BRD4 action. Studies on inhibitors have inherent limitations owing to uncertainty regarding their specificity, modality of action, and long-term consequences. For example, the impact of BET inhibitors on normal hematopoietic cells is not well understood, posing potential problems when treating blood cancers such as leukemia/lymphoma. BET inhibitor treatment may compromise the activity and maintenance of hematopoietic stem cells and may weaken the ability to combat infection, which is also relevant to treating inflammation, given that macrophages are the main effector of both inflammation and host defense.
We thus sought to gain a fuller understanding of BRD4’s activity in normal hematopoiesis and during inflammatory and innate immune responses. We studied Brd4 conditional knockout mice, focusing on hematopoiesis and macrophage responses. First, we tested mice in which Brd4 is deleted in early hematopoiesis by using the Vav-Cre technique. We showed that Brd4 KO mice die during fetal development owing to severe defects in the expansion of hematopoietic stem cells (HSC) and in the development of hematopoietic progenitor cells. As a consequence, Brd4 KO embryos fail to develop immune cells of all lineages, including lymphocytes and myeloid cells, which are important for innate and adaptive immunity. We also found that BRD4 is essential for the proliferation of macrophages, based on LysM-Cre–dependent deletion of Brd4 (LysM-Cre selectively targets macrophages and neutrophils); the resultant Brd4 KO mice failed to start IL-4–dependent peritoneal macrophage expansion. The results strongly point to a central role of BRD4 in immune cell expansion, required for maintaining immunity.
We investigated the genome-wide distribution of BRD4 in macrophages in a resting condition and after LPS stimulation. LPS is a pathogen component that rapidly induces inflammatory genes and interferon-stimulated genes important for protection against pathogens. We found that BRD4 broadly occupies genic and intergenic regions. Within the genic region, BRD4 binding peaked at the transcription start site (TSS), although binding was detected over the 5′ promoter and within the coding regions. BRD4 binding over the genic regions markedly increased after LPS stimulation, indicating that BRD4 moves rapidly over the genome, presumably to accommodate a rapid alteration of histone acetylation. Furthermore, BRD4 displayed dense clustering over distant regulatory regions that represented superenhancers. BRD4 clusters coincided with the H3K27 chromatin mark, which denotes superenhancers as well as RNA polymerase II clustering. BRD4–containing superenhancers localized to genes important for basic macrophage phenotypes and innate immune responses. We also showed that superenhancers rapidly redistributed in response to LPS in WT macrophages. Interestingly, superenhancers were found even in Brd4 KO macrophages. These superenhancers contained large clusters of pol II and H3K27ac marks, but without BRD4, and their distribution patterns were anomalous. The results led us to conclude that BRD4 plays an important role in shaping superenhancers necessary for inflammatory responses in macrophages.
Additional Funding
- NICHD Scientific Director's Award (2018–2019)
- Washington University RVCL (Retinal Vasculopathy with Cerebral Leukodystrophy) Foundation
- Washington University RVCL Foundation
Publications
- Kamada R, Yang W, Xhang Y, Patel M, Yang Y, Wakabayashi Y, Ouda R, Sakagichi K, Fujita T, Tamura T, Zhu, J, Ozato K. Interferon stimulation creates heritable chromatin marks to establish transcriptional memory. Proc Natl Acad Sci USA 2018;115(39):E9162-E9171.
- Bachu M, Tamura T, Chen C, Narain A, Nehru V, Sarai N, Ghosh SB, Ghosh A, Kavarthapu R, Dufau ML, Ozato K. A versatile mouse model of epitope-tagged histone H3.3 to study epigenome dynamics. J Biol Chem 2019;294:1904-1914.
- Dey A, Yang W, Gegonne A, Nishiyama A, Pan R, Yagi R, Grinberg A, Finkelman FD, Pfeifer K, Zhu J, Singer D, Zhu J, Ozato K. BRD4 directs hematopoietic stem cell development and modulates macrophage inflammatory responses. EMBO J 2019;38(7):e100293.
- Gegonne A, Chen QR, Dey A, Etzensperger R, Tai X, Singer A, Meerzaman D, Ozato K, Singer DS. Immature CDS single-positive thymocytes are a molecularly distinct subpopulation, selectively dependent on BRD4 for their differentiation. Cell Rep 2018;24:117-129.
- Thumbigere-Math V, Foster BL, Bachu M, Yoshii H, Brooks SR, Coulter A, Chavez MB, Togi S, Neely AL, Deng Z, Mansky KC, Ozato K, Somerman MJ. Inactivating mutation in IRF8 promotes osteoclast transcriptional programs and increases susceptibility to tooth root resorption. J Bone Miner Res 2019;34(6):1155-1168.
Collaborators
- Steven L. Coon, PhD, Molecular Genomics Core, NICHD, Bethesda, MD
- Robert J. Crouch, PhD, Section on Formation of RNA, NICHD, Bethesda, MD
- Herbert Morse II, MD, Laboratory of Immunopathology, NIAID, Rockville, MD
- Yoh-suke Mukouyama, PhD, Laboratory of Stem Cell and Neuro-Vascular Biology, NHLBI, Bethesda, MD
- Dinah S. Singer, PhD, Experimental Immunology Branch, NCI, Bethesda, MD
- Tomohiko Tamura, MD, PhD, Tokyo University, Tokyo, Japan
- Jun Zhu, PhD, DNA Sequencing and Genomics Core, NHLBI, Bethesda, MD
Contact
For more information, email ozatok@mail.nih.gov or visit http://ozatolab.nichd.nih.gov.


