Chromatin Remodeling and Gene Activation

- David J. Clark, PhD, Head, Section on Chromatin and Gene Expression
- Peter Eriksson, PhD, Staff Scientist
- Razvan V. Chereji, PhD, Research Fellow
- Christopher Coey, PhD, Intramural Research Training Award Fellow
- Hemant K. Prajapati, PhD, Visiting Fellow
- Allison F. Dennis, BS, Graduate Student
- LauraAnn Schmidberger, BS, Postbaccalaureate Fellow
Our primary goal is to understand how chromatin structure influences gene regulation. Chromatin is generally repressive in nature, but its structure is manipulated by cells in a regulated way to determine which genes are potentially transcriptionally active and which genes remain repressed in a given cell type. Such regulation depends on interactions between DNA sequence–specific transcription factors, chromatin enzymes, and chromatin.
The structural subunit of chromatin is the nucleosome core, which contains 147 bp of DNA wrapped 1.7 times around a central histone octamer; the octamer is composed of two molecules each of the four core histones (H2A, H2B, H3, and H4). Generally, nucleosomes are regularly spaced along the DNA, like beads on a string. At physiological salt concentrations, the beads-on-a-string structure folds spontaneously to form a 30 nm–wide fiber, assisted by the linker histone (H1), which binds to the nucleosome core and to the linker DNA. Collectively, the histones thus determine DNA accessibility.
Gene activation involves the recruitment of a set of factors to a promoter in response to appropriate signals, ultimately resulting in the formation of an initiation complex by RNA polymerase II (Pol II) and in transcription. For transcription to occur, the promoter must be cleared of nucleosomes to allow transcription complex formation, and then Pol II must negotiate the nucleosomes on the gene. Nucleosomes are compact structures capable of blocking transcription at every step. To circumvent and regulate such chromatin blocks, eukaryotic cells possess dedicated enzymes, including ATP–dependent chromatin remodeling machines, histone-modifying complexes, and histone chaperones. The remodeling machines use ATP to move nucleosomes along or off DNA (e.g., the SWI/SNF, RSC, CHD, and ISWI complexes), or to exchange histone variants between nucleosomes (e.g., the SWR complex). The histone-modifying complexes contain enzymes that modify the histones posttranslationally to alter their DNA–binding properties and to mark them for recognition by other complexes, which have activating or repressive roles (the "histone code" hypothesis). Histone-modifying enzymes include histone acetylases (HATs), deacetylases (HDACs), methylases, and kinases. Histone chaperones mediate histone transfer reactions that occur during transcription and DNA replication (e.g., FACT, Asf1, and the CAF-1 complex). These enzymes, together with the DNA–methylating and –demethylating enzymes present in higher eukaryotes, are central to epigenetics.
Many human diseases have been linked to chromatin-remodeling enzymes and epigenetic modifications. For example, mutations in the hSNF5 subunit of the SWI/SNF complex are strongly linked to pediatric rhabdoid tumors. The CHD class of ATP–dependent remodelers has also been linked to cancer and to autism. Cancer therapies and drugs aimed at epigenetic targets are being tested. Recent studies have revealed a correlation between a linker histone variant and tumor heterogeneity. A full understanding of chromatin structure and the mechanisms by which it is manipulated is therefore of vital importance.
Interplay between the RSC, ISW1, CHD1, and ISW2 chromatin-remodeling complexes
Our aim is to dissect chromatin remodeling mechanisms in vivo and to understand their contributions to gene regulation. Our current efforts focus on elucidating the contributions of the various ATP–dependent remodeling complexes to chromatin organization in vivo. During this Fiscal Year, we made significant progress towards understanding the roles of the RSC (remodeling the structure of chromatin) ATP–dependent remodeling complex in gene regulation. RSC is an essential SWI/SNF–like remodeler that is similar to the mammalian PBAF chromatin-remodeling complex and plays an important role in determining the size of the nucleosome-depleted region (NDR) typically found at potentially active gene promoters. In both mammals and yeast, promoter NDRs are flanked by arrays of regularly spaced nucleosomes, which are phased relative to the transcription start site.
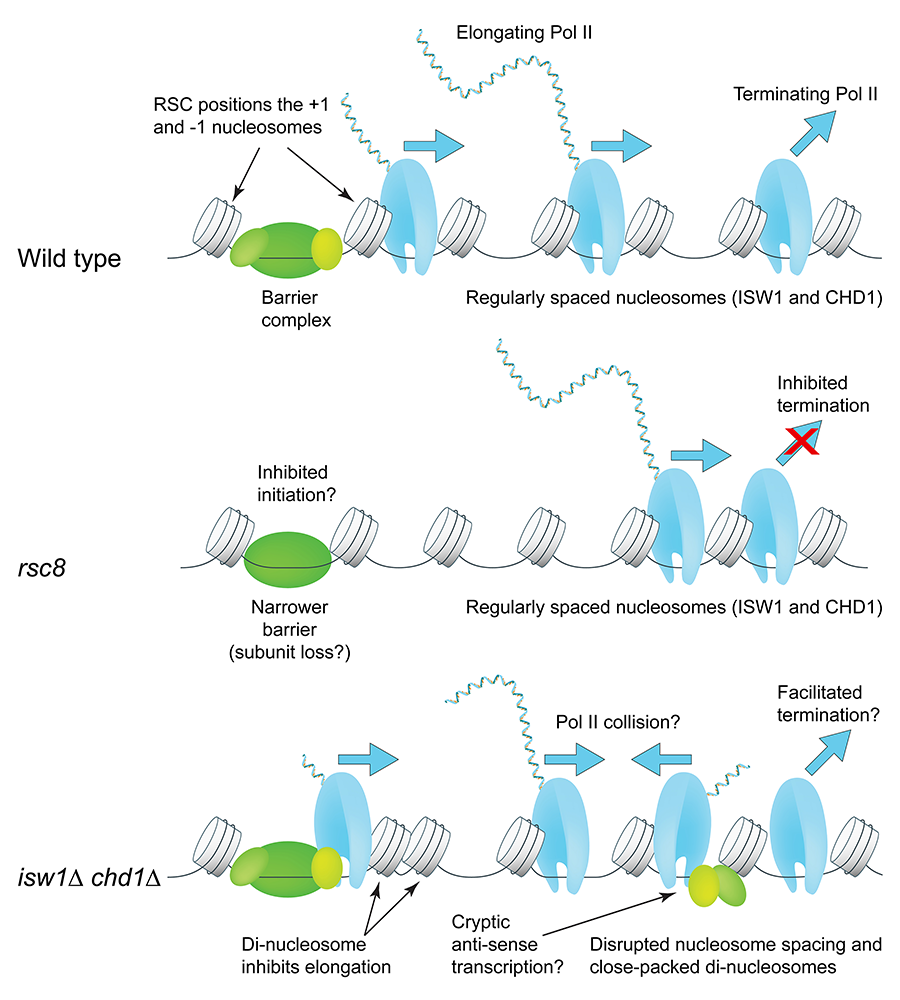
We examined the interplay between RSC and the ISW1, CHD1, and ISW2 ATP–dependent nucleosome-spacing enzymes in chromatin organization and transcription, using isogenic yeast strains lacking all combinations of these enzymes [Reference 1]. The contributions of these remodelers to chromatin organization are largely combinatorial, distinct, and non-redundant, supporting a model in which the first (+1) nucleosome on the gene is positioned by RSC and then used as a reference nucleosome by the spacing enzymes to create phased nucleosomal arrays. We observed that defective chromatin organization correlates with altered Pol II distribution: RSC–depleted cells exhibit lower levels of elongating Pol II and higher levels of terminating Pol II, consistent with defects in both termination and initiation, suggesting that RSC facilitates both. Cells lacking both ISW1 and CHD1 show the opposite Pol II distribution, suggesting opposite elongation and termination defects. Such cells have extremely disrupted chromatin, with high levels of closely packed dinucleosomes (i.e., two nucleosomes with no intervening linker DNA), primarily involving the second (+2) nucleosome on the gene (i.e., dinucleosomes composed of either the +1 and +2 nucleosomes or the +2 and +3 nucleosomes). We propose that ISW1 and CHD1 facilitate Pol II elongation by separating nucleosomes that are too close together (Figure 1).
Click image to view.
Figure 1. Roles of the RSC, ISW1, and CHD1 ATP–dependent chromatin remodelers in RNA polymerase II initiation, elongation and termination
RSC widens the nucleosome-depleted region at the promoter by repositioning the flanking +1 and –1 nucleosomes farther apart to accommodate a complete barrier complex. CHD1 (short spacing) and ISW1 (longer spacing) compete to set the nucleosome spacing on a gene using the +1 nucleosome as a reference. In cells lacking RSC (rsc8), nucleosomes shift toward the promoter, but the spacing is the same as in wild-type cells; Pol II lingers 3′ of the transcript-termination site. In the isw1 chd1 double mutant, nucleosome spacing is disrupted, with the +2 nucleosome in particular being pushed against the +1 or +3 nucleosome. High cryptic initiation in these cells may result in collisions between transcribing cryptic anti-sense Pol II and promoter-initiated Pol II (elongation defect). From Reference 1.
Role of the RSC ATP–dependent chromatin remodeling complex in transcription factor dynamics
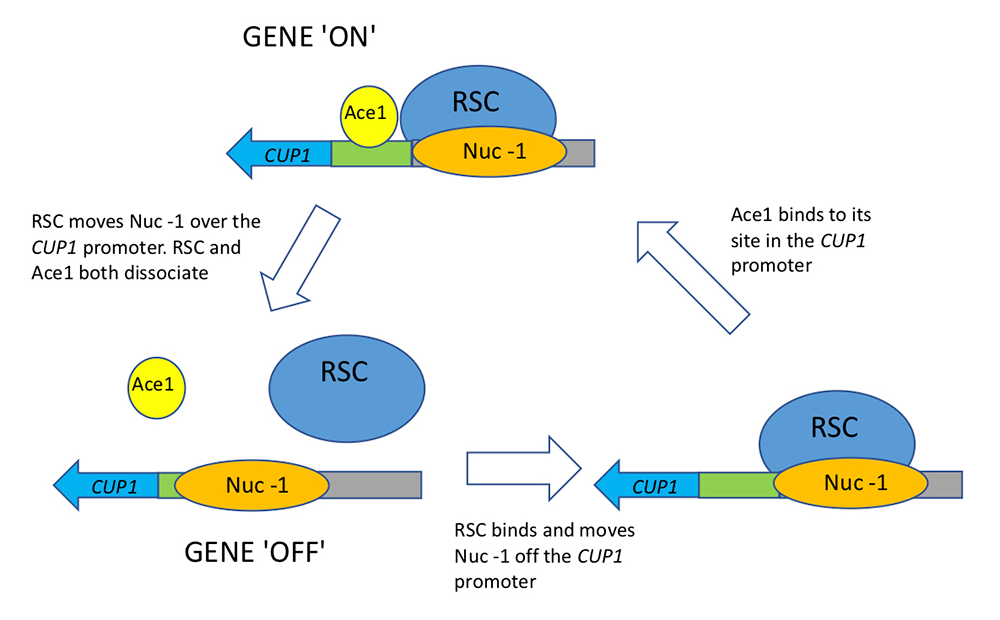
In a collaboration with the Karpova lab, we investigated the role of RSC in transcription factor dynamics [Reference 2]. Generally, the binding of sequence-specific transcription factors to cognate sites is highly dynamic. However, how such binding is linked to chromatin remodeling and transcription is unclear. The CUP1 gene encodes a metallothionein responsible for protecting cells against the toxic effects of copper ions; it is induced when excess copper ions bind to the Ace1p transcription factor, which then binds to upstream activating sequences (UAS elements) in the CUP1 promoter to activate transcription. Using single-molecule tracking, we showed that RSC reduces the time it takes for the Ace1p transcription factor to locate and bind to the CUP1 promoter. We quantified smFISH (single molecule–fluorescence in situ hybridization) mRNA data using a gene-bursting model and demonstrated that RSC regulates CUP1 transcription bursts by modulating Ace1p transcription factor occupancy, rather than by affecting initiation and elongation rates. The single-molecule tracking data show that RSC binds to the activated CUP1 promoter transiently. We also observed that RSC does not affect nucleosome occupancy at CUP1. We propose that transient binding of Ace1p and rapid bursts of CUP1 transcription may depend on short, repetitive cycles of RSC–mediated nucleosome mobilization (Figure 2). This type of regulation would reduce transcriptional noise and ensure a homogeneous response of the cell population to copper stress.
Click image to view.
Figure 2. Linked cycles of RSC chromatin remodeling and Ace1 transcription factor binding in yeast
Single-molecule tracking and smFISH reveal the link between chromatin remodeling by the RSC complex and Ace1 transcription factor binding. Cycles of Ace1 binding at the CUP1 promoter lead to bursts of CUP1 transcription, which depend on short cycles of nucleosome mobilization by RSC (the –1 nucleosome on the promoter). RSC ensures a homogeneous response of the cell population to copper-mediated oxidative stress. From Reference 2.
Role of tRNA genes in chromosome organization in yeast
We also collaborated with the Kamakaka lab to investigate the role of tRNA genes in chromosomal organization in the nucleus [Reference 3]. The genome is packaged and organized in an ordered, nonrandom manner within the nucleus, involving contacts between specific chromatin segments and nuclear substructures. tRNA genes are binding sites for transcription factors as well as for architectural proteins and are thought to play an important role in such organization. We tested the hypothesis by removing all tRNA genes from chromosome III, either by deletion or by transfer to another chromosome. Surprisingly, loss of all tRNA genes does not grossly affect chromatin architecture or chromosome tethering and mobility. However, loss of tRNA genes does affect local chromatin structure by altering both nucleosome positioning and the binding of proteins involved in maintaining chromosome architecture (cohesins and condensins), as expected. The absence of tRNA genes also alters centromere clustering and reduces the frequency of long-range heterochromatin clustering, with concomitant effects on gene silencing.
Publications
- Ocampo J, Chereji RV, Eriksson PR, Clark DJ. Contrasting roles of the RSC and ISW1/CHD1 chromatin remodelers in RNA polymerase II elongation and termination. Genome Res 2019;29:407-417.
- Mehta GD, Ball DA, Eriksson PR, Chereji RV, Clark DJ, McNally JG, Karpova TS. Single-molecule analysis reveals linked cycles of RSC chromatin remodeling and Ace1p transcription factor binding in yeast. Mol Cell 2018;72:875-887.
- Hamdani O, Dhillon N, Hsieh TS, Fujita T, Ocampo J, Kirkland JG, Lawrimore J, Kobayashi TJ, Friedman B, Fulton D, Wu KY, Chereji RV, Oki M, Bloom K, Clark DJ, Rando OJ, Kamakaka RT. tRNA genes affect chromosome structure and function via local effects. Mol Cell Biol 2019;39:e00432-18.
Collaborators
- Munira A. Basrai, PhD, Genetics Branch, Center for Cancer Research, NCI, Bethesda, MD
- Harold Burgess, PhD, Section on Behavioral Neurogenetics, NICHD, Bethesda, MD
- Douglas Fields, PhD, Section on Nervous System Development and Plasticity, NICHD, Bethesda, MD
- Chhabi Govind, PhD, Oakland University, Rochester, MI
- Gordon L. Hager, PhD, Laboratory of Receptor Biology and Gene Expression, Center for Cancer Research, NCI, Bethesda, MD
- Jeffrey J. Hayes, PhD, University of Rochester Medical Center, Rochester, NY
- Alan G. Hinnebusch, PhD, Section on Nutrient Control of Gene Expression, NICHD, Bethesda, MD
- Rohinton T. Kamakaka, PhD, University of California Santa Cruz, Santa Cruz, CA
- Tatiana Karpova, PhD, Laboratory of Receptor Biology and Gene Expression, Center for Cancer Research, NCI, Bethesda, MD
- Philip R. Lee, PhD, Section on Nervous System Development and Plasticity, NICHD, Bethesda, MD
- Vasily M. Studitsky, PhD, Fox Chase Cancer Center, Philadelphia, PA
Contact
For more information, email clarkda@mail.nih.gov or visit http://clarklab.nichd.nih.gov.


