Mechanisms of Nuclear Genome Organization and Maintenance

- Mary Dasso, PhD, Head, Section on Cell Cycle Regulation
- Vasilisa Aksenova, PhD, Visiting Fellow
- Alexei Arnaoutov, PhD, Staff Scientist
- Maia Ouspenskaia, DVM, Biologist
- Shane Chen, PhD, Postdoctoral Intramural Research Training Award Fellow
- Karen M. Plevock Haase, PhD, Postdoctoral Intramural Research Training Award Fellow
- Saroj Regmi, PhD, Postdoctoral Intramural Research Training Award Fellow
- Sophie Bice, BA, Postbaccalaureate Intramural Research Training Award Fellow
- Abigail Daniels, Postbaccalaureate Intramural Research Training Award Fellow
- Ross T. Kaufhold, BA, Postbaccalaureate Intramural Research Training Award Fellow
- Ka Chun Yau, BS, Predoctoral Visiting Fellow
- Hang Noh Lee, PhD, Research Fellow
We are interested in mechanisms of genome maintenance and organization. During interphase, chromosomes are surrounded by the nuclear envelope (NE), which separates the nuclear and cytoplasmic compartments of the cell. The sequestration of chromosomes within the nucleus has profound consequences for almost all aspects of gene expression and cell function. Communication between the nucleus and cytoplasm occurs through conduits called nuclear pore complexes (NPCs), which are embedded in the NE and consist of about 30 proteins called nucleoporins (Figure 1). Beyond nucleocytoplasmic trafficking, nucleoporins are important for chromosome organization, transcriptional control, RNA processing, cell signaling, and cell-cycle control. Both nucleoporins and soluble components of the nuclear trafficking machinery also perform transport-independent functions in mitotic chromosome segregation. The involvement of nucleoporins in such diverse events offers the intriguing possibility that they might coordinate these processes with nuclear trafficking and with each other. Moreover, nucleoporin dysfunction has important clinical implications; nucleoporin genes are frequently misregulated in cancers, and nucleoporin mutations cause congenital defects, pediatric nephrotic syndromes, and premature ovarian insufficiency. Nucleoporins are critical viral targets, and their disruption contributes to neurodegenerative conditions, including amyotrophic lateral sclerosis, frontotemporal dementia, and Huntington’s disease.
Our research studies nucleoporins, NPC–associated proteins (e.g., the SUMO pathway, spindle checkpoint proteins), and other components of the nuclear transport machinery (e.g., the Ran pathway) throughout the cell cycle. Our goal is to define their biochemical roles and how their dysregulation causes human disease. We have taken a multifaceted approach toward this question, using both CRISPR–based strategies in mammalian cells and Drosophila developmental genetics.
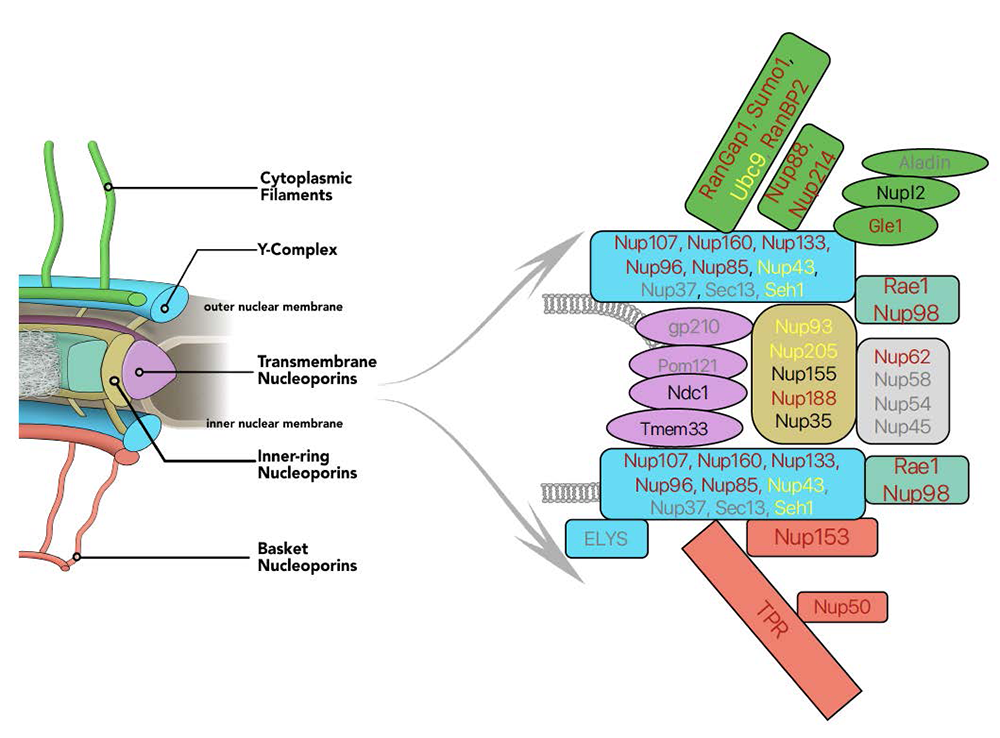
Click image to view.
Figure 1. AID tagging of nucleoporins
Left side shows model of nuclear pore structure with cytoplasmic fibers (green) oriented upward and nucleoplasmic basket (red) oriented downward. Other domains of the NPC as indicated. Right side represents the polypeptides associated with NPC sub-complexes, with fill colors corresponding to model domains. Text shows progress in obtaining AID–tagged cell lines for individual nucleoporins: red: successful tagging; black: in progress; yellow: tagging achieved but cell line quality issues; grey: tagging unsuccessfully attempted.
Selective degradation of nucleocytoplasmic transport proteins and their interphase function
Understanding the activities of individual nucleoporins has been complicated by their multifaceted nature, abundance, and unusual stability. To better address this question, we employed strategies for selective and rapid degradation of individual proteins within human tissue-culture cells (Figure 2). Specifically, we used CRISPR/Cas9 to construct cell lines in which sequences encoding auxin-induced degron (AID) domains are inserted into both alleles of targeted genes within cells that also stably express the Transport Inhibitor Response 1 (TIR1) protein. TIR1 acts as a subunit of the SCF ubiquitin ligase complex, so that the AID–tagged fusion proteins undergo rapid, selective degradation upon addition of the plant hormone auxin. We frequently also add a fluorescent tag to the targeted proteins, allowing their degradation to be monitored visually as well as biochemically. We have been successful in developing cell lines that allow conditional depletions of nucleoporins associated with different regions of the NPC (Figure 1).
Our current studies regarding the roles of nucleoporins during interphase address three issues. First, we are examining the role of individual nucleoporins in NPC assembly and stability. Our results indicate that different regions of the NPC can persist independently after depletion of individual nucleoporins, suggesting that the NPC is a surprisingly modular structure. Second, we are examining the role of individual nucleoporins in different nuclear trafficking pathways, an assessment that includes evaluation of nuclear protein import, protein export, and RNA export. We find distinct sensitivities of these pathways to the loss of individual nucleoporins, supporting the idea that the transport pathways have distinct NPC structural requirements. Third, we are examining gene expression by RNA-seq with and without auxin, to observe the impact of acute or prolonged nucleoporin depletion. Our results suggest that particular nucleoporins have distinct and nonredundant roles in gene expression. Defining which individual nucleoporins contribute to each of these processes will allow us to better design future experiments examining the role of those selected proteins in the regulation of cellular events.
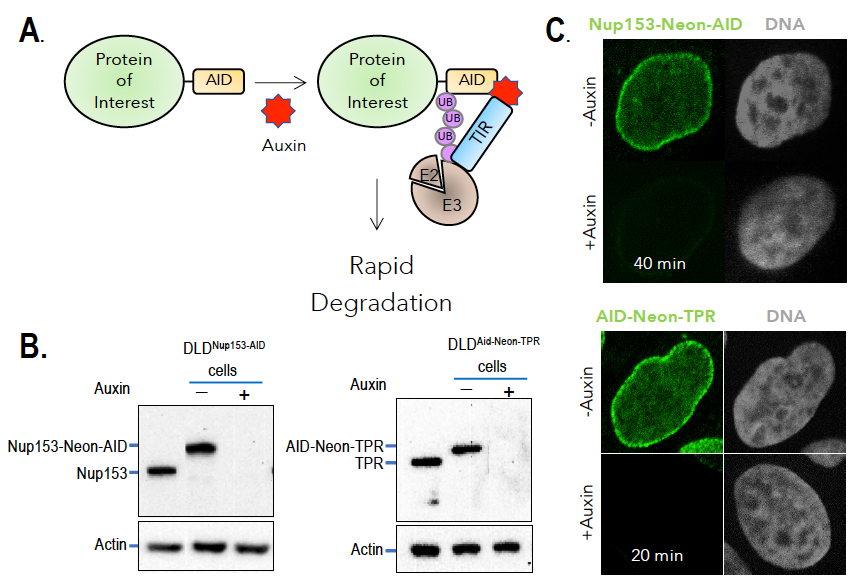
Click image to view.
Figure 2. Auxin-induced degradation of AID–tagged nucleoporins
A. TIR1–expressing cells ubiquitinate and degrade proteins tagged with auxin-induced degron (AID) domains upon auxin addition. Nup153 and TPR were homozygously fused with the AID tag and a fluorescent marker (neon Green) in TIR1–expressing cells. Rapid and uniform degradation is observed after auxin addition by Western blotting (B) or destruction of the fluorescent tag (C).
Roles of nucleoporins in gene expression and RNA trafficking
One example of how our studies have addressed important questions in the nucleoporin field is our findings regarding the NPC nuclear basket. The vertebrate NPC nuclear basket consists of three basket nucleoporins (BSK-NUPs) called Nup153, Tpr, and Nup50. Beyond trafficking, BSK-NUPs have been implicated in chromatin remodeling, control of gene expression and protein modification, and mRNA processing. To understand the role of BSK-NUPs in these processes, we analyzed AID–tagged cell lines for each of these nucleoporins. We found that Nup153 and Tpr each remain associated with NPCs in the absence of the other, but that Nup50 localization depends on Nup153. No other nucleoporins showed significant changes in localization after depletion of any BSK-NUP. Loss of each nucleoporin caused unique transport and transcriptomic changes, validating our central hypothesis that individual nucleoporins play distinct and specific roles. Notably, not only were transcriptomic changes distinct among BSK-NUPs, but their profiles also differed from changes after depletion of RanGAP1 and the loss of Ran–mediated nuclear trafficking.
We further examined the role of Tpr in gene expression, because it showed the most rapid and significant gene expression changes. GANP is a scaffolding subunit of the TRanscription and EXport 2 (TREX-2) complex, which bridges the transcription and export machineries in yeast through association with the Mediator complex and the NPC. We observed that the GANP protein associated with the NPC in a Tpr–dependent manner, suggesting a possible link between Tpr–dependent transcriptomic changes and altered RNA export. We analyzed cell lines in which GANP or other key RNA export factors were AID–tagged and observed changes in RNA abundance closely related to changes after Tpr depletion (Figure 3), suggesting an integral role of Tpr in TREX-2–dependent RNA export. Both up- and downregulated transcripts showed retention within nuclei in the absence of either Tpr or GANP, further illustrating a critical role of Tpr in TREX-2–mediated mRNA export. Our ongoing experiments are investigating the mechanisms through which Tpr facilitates TREX-2–dependent mRNA export.
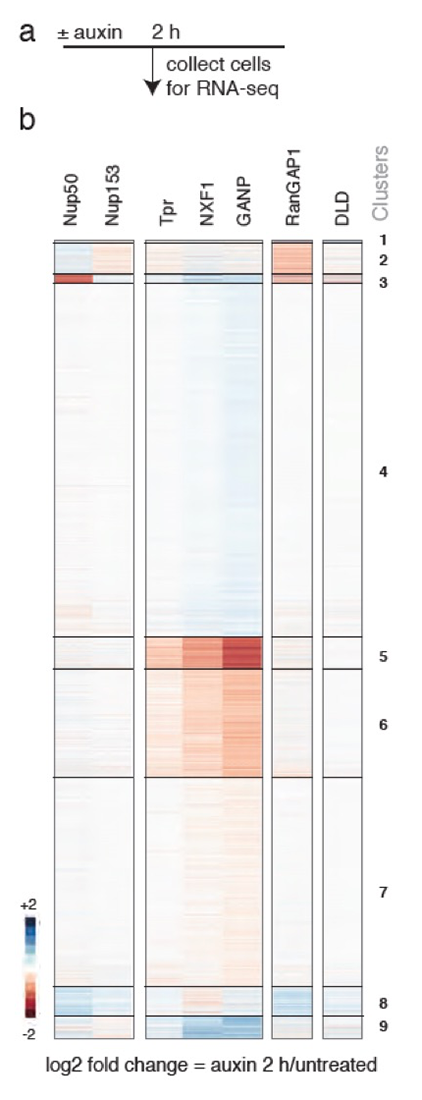
Click image to view.
Figure 3. Loss of Tpr or TREX-2 (DGANP) causes similar rapid changes in mRNA abundance.
Heatmaps of unsupervised k-means clustering of differentially expressed genes 2 h after auxin treatment of cells expressing corresponding AID–tagged proteins. DLD: parental DLD1 cells treated with auxin. DNXF1 shows a broader arrest of RNA trafficking, and DRanGAP1 shows arrest of Ran–dependent transport. Note the distinct patterns after depletion of other BSK–NUPs, Nup153, and Nup50.
Mitotic roles of the nuclear trafficking machinery
We have a long-standing interest in the process of chromosome segregation. Defects in chromosome segregation lead to aneuploidy, a condition in which cells possess an abnormal number of chromosomes. Several common birth defects, such as Down's syndrome, result from aneuploidy arising during meiotic cell divisions, and aneuploidy arising from mitotic divisions is a hallmark of many types of solid tumors. The Ran GTPase pathway and nucleoporins promote chromosome segregation through their important roles in spindle assembly and cell-cycle progression. To segregate chromosomes accurately, Ran–GTP distribution must be tightly regulated, both spatially and temporally. Ran’s cytoplasmic GTPase–activating protein, RanGAP1, and its chromatin-bound nucleotide exchange factor, RCC1, establish an interphase distribution, with GTP–bound Ran (Ran-GTP) in the nucleus and GDP–bound Ran (Ran-GDP) in the cytosol. Such asymmetry drives nucleocytoplasmic trafficking through controlled cargo binding and release by karyopherins, a family of Ran-GTP–binding transport receptors. After mitotic NE breakdown (NEB), RCC1 generates Ran-GTP near chromosomes, while Ran distal to chromosomes is GDP–bound, directing spindle assembly through spatially regulated release of spindle assembly factors (SAFs) from karyopherins. Notably, the error-prone nature of Ran–dependent meiotic spindle assembly may contribute to human pregnancy losses and genetic disorders, including Down's syndrome. RanBP1 is a Ran-GTP–binding protein that forms a stable heterotrimeric complex with Ran and RCC1 in vitro (RRR complex), inhibiting RCC1’s nucleotide exchange activity.
Xenopus egg extracts (XEEs) are a well-established ex vivo model system for mitotic studies. XEEs possess large amounts of RCC1, which is stockpiled in eggs to facilitate early development. We previously reported that RRR complex formation in XEEs determines RCC1’s partitioning between its chromatin-bound and soluble forms and inhibits the exchange activity of soluble RCC1. We found that dissolution of the RRR complex at anaphase onset creates a large pool of free RCC1, which becomes available for chromatin binding. We further investigated how RRR–complex dynamics influences kinetochore and centromere function within XEE. Kinetochores are proteinaceous structures that act as attachment sites for spindle microtubules. Kinetochores assemble during each mitosis on centromeres, specialized chromatin domains that are present at a single locus on each of the sister chromatids. Centromeres are also sites of chromatid cohesion, which is maintained until anaphase onset, when centromeres must be rapidly and synchronously re-modeled to release sister chromatid cohesion and thus allow chromosome segregation into daughter cells and cytokinesis. We studied replicated chromosomes in XEE that possess physiologically paired sister chromatids and found that mimicking the anaphase wave of free RCC1 by adding exogenous RCC1 causes the specific release of chromatid cohesion. Strikingly, RCC1 mutants that lacked activity as a Ran exchange factor were as effective wild-type RCC1 in this assay, whereas active but chromatin binding–deficient RCC1 mutants or Ran-GTP itself were not active. Together, these data indicate that RCC1 can regulate anaphase kinetochore composition in a Ran-GTP–independent fashion; this is the first report of a biological function of RCC1 that is independent of its activity as a Ran exchange factor.
We recently investigated the relevance of the RRR complex to mitosis in somatic cells. Most RCC1 localizes on or near chromosomes throughout mitosis in non-embryonic systems, which have relatively low levels of soluble RCC1, raising questions about whether RRR complex formation significantly controls RCC1 dynamics after early development. Nevertheless, RanBP1 depletion by RNAi disrupts mitotic progression in mammalian tissue-culture cells. Notably, the dynamics for chromosome-bound RCC1 are not uniform as tissue culture cells progress through mitosis. To better understand mitotic RCC1 dynamics, we investigated whether RanBP1 or a related protein called RanBP3 might be important for controlling mitotic Ran-GTP gradients within somatic cells. To address this question, we systematically varied RanBP1 and RanBP3 levels in HCT116 or DLD1 cell lines through overexpression or fusion with AID tags. Consistent with earlier reports, we found that RanBP1 was dispensable for interphase import or export of a model substrate, while RanBP3 appears to facilitate nuclear export via the Crm1 karyopherin. Within mitosis, we observed that altering RanBP1 concentrations substantially altered RCC1 dynamics on metaphase chromosomes, while altering RanBP3 did not. Moreover, we found dramatic re-localization of the SAF Hepatoma Up-Regulated Protein (HURP, a component of the spindle-assembly pathway) during metaphase in direct correspondence with changes in RCC1 dynamics (Figure 4), showing that Ran-GTP levels and SAF activity near chromosomes correlate with altered RCC1 behavior. Analogous to findings in XEE, these data indicate an important mitotic role in human somatic cells for RanBP1 in controlling RCC1 dynamics and determining the accurate the spatial distribution and magnitude of the Ran-GTP gradients, thus ensuring correct execution of Ran–dependent mitotic events.
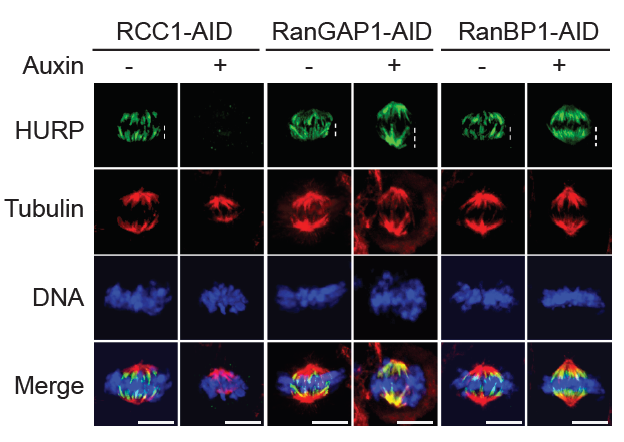
Click image to view.
Figure 4. Regulation of mitotic Ran–GTP gradients by RRR complex
Immuno-staining with HURP and Tubulin antibodies of cells in HCT116RCC1-µAID-3xFLAG, HCT116RanGAP1-3mAID, and HCT116RanBP1-µAID-HA cells treated with or without 1 mM Auxin for 5, 2, and 3 h, respectively. Cells also express TIR1. White dashed lines represent the length of HURP signal (top row). Scale bars = 10 μm.
The role of the IRBIT protein in tissue homeostasis
We previously reported a conserved role for the IRBIT protein (IP3-receptor-binding protein released with inositol 1,4,5-trisphosphate) in inhibiting ribonucleotide reductase (RNR), an enzyme that produces deoxynucleotide triphosphates (dNTPs) within the cell for DNA synthesis. We further found that mammalian tissue-culture cells show altered cell-cycle progression and genome stability that was potentially disrupted in the absence of IRBIT, and that this mechanism is conserved between humans and flies (Drosophila melanogaster). Therefore, in collaboration with Mihaela Serpe and Brian Oliver, we chose to use flies as a model organism to understand the role of this mechanism in development and tissue homeostasis.
In situ hybridization shows IRBIT expression in regions destined to become the midgut during embryogenesis, and IRBIT is highly expressed in the adult midgut. The Drosophila midgut has a tubular structure and is surrounded by visceral muscles. The adult midgut possesses a monolayered epithelium that is composed of four distinct cell types (Figure 5B): intestinal stem cells (ISCs), undifferentiated progenitor cells called enteroblasts (EBs), specialized absorptive enterocytes (ECs), and secretory enteroendocrine cells (EEs). The midgut is maintained through division of ISCs, giving rise to EBs, which in turn differentiate into EEs. Nutrients are absorbed from the lumen of the gut, which also contains a complex microbiota; the midgut acts both as a niche for commensal microbes and as the first line of defense for against microbial pathogens. Like the intestine of vertebrates, the epithelium of the midgut has a remarkable regenerative capacity, which has been extensively exploited for the study of stem cell–driven tissue self-renewal, as well as tissue homeostasis during aging.
We examined IRBIT’s potential role in the midgut by generating an IRBIT null fly (IRBIT–/–) (Figure 5A). The midguts of one-day-old wild-type and IRBIT–/– flies were essentially indistinguishable at the tissue-architecture level. However, we observed a rapid loss of tissue homeostasis in the IRBIT–/– flies, with a progressive increase in relative numbers of undifferentiated enteroblast progenitor cells and tissue dysplasia. IRBIT–/– flies also show fewer cell-cell contacts when stained for junctional proteins in the posterior midgut epithelium and altered gene expression patterns, reminiscent of changes associate with inflammation and aging. The phenotypes are fully rescued through expression of full-length IRBIT, and further experiments suggested that altered dNTP pools likely contribute to the IRBIT–/– phenotypes. Further analysis showed that the IRBIT–RNR pathway is essential to ensure correct differentiation of intestinal stem cell (ISC) progeny, and that it is a key downstream target of the GATAe transcription factor. Moreover, the GATAe–IRBIT–RNR pathway becomes dysfunctional as flies age, contributing to a characteristic accumulation of undifferentiated ISC progeny that can be reversed by specifically inhibiting RNR within progeny cells. Collectively, our findings showed that RNR suppression by IRBIT is an important mechanism that directs differentiation of ISC progeny to maintain intestinal tissue homeostasis.
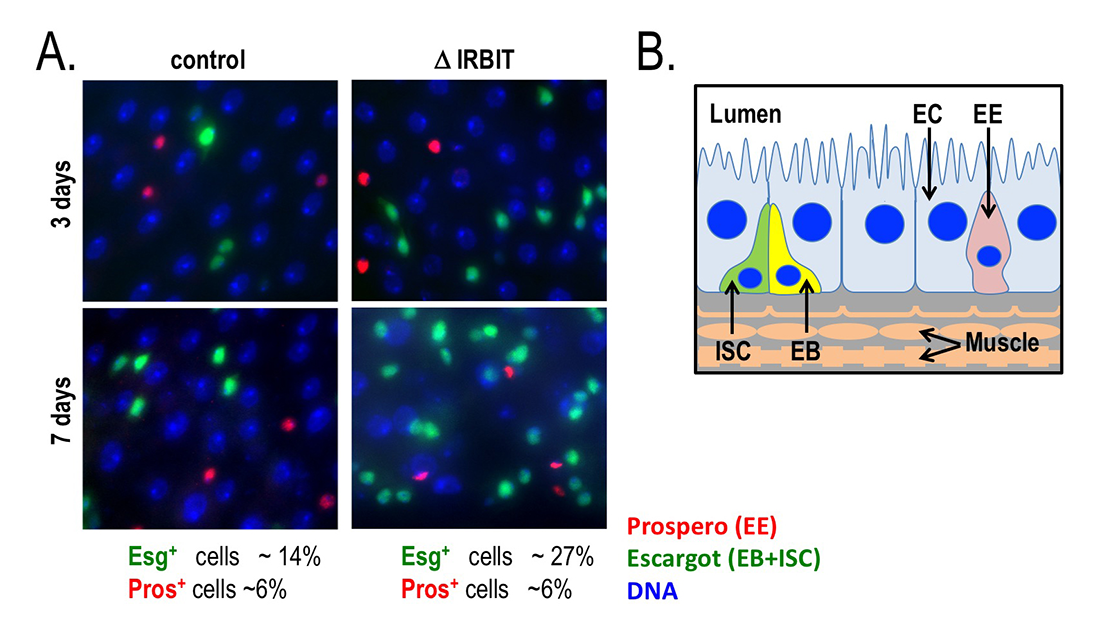
Click image to view.
Figure 5. Loss of IRBIT disrupts tissue homeostasis in the Drosophila midgut.
A. Epithelia from control (left) and IRBIT–/– (right) flies at three (top) or seven (bottom) days after eclosure. The guts were stained with DNA dye Hoechst 33258 (blue), and antibodies against Prospero (EE cells) and Escargot (EB and ISC cells). We observed a rapid and progressive increase in the fraction of Escargot-positive cells in the IRBIT–/– flies over time. In conjunction with additional experiments, the accumulation is indicative of accumulation of undifferentiated enteroblast progenitor cells.
B. Schematic of epithelium within the Drosophila midgut. EC: enterocyte; ISC: intestinal stem cell; EB: enteroblast; EE: enteroendocrine cell.
Additional Funding
- NICHD DIR Director’s Investigator Award
- NICHD Intramural Research Fellowship (Saroj Regmi)
Publications
- Aksenova V, Lee HN, Smith A, Chen S, Bhat P, Iben J, Echeverria C, Fontoura B, Arnaoutov A, Dasso M. Distinct basket nucleoporins' roles in nuclear pore function and gene expression: Tpr is an integral component of the TREX-2 mRNA export pathway. bioRxiv 2019;685263.
- Arnaoutov A, Lee HN, Plevock Haase K, Aksenova V, Jarnik M, Oliver B, Serpe M, Dasso M. IRBIT directs differentiation of intestinal stem cell progeny to maintain tissue homeostasis. bioRxiv 2019;737262.
- Hassebroek V, Pandey N, Park H, Aksenova V, Arnaoutouv A, Dasso M, Azuma Y. Mitotic SUMOylation contributes to PICH and topoisomerase IIa cooperation by promoting their interaction on chromosomes. bioRxiv 2019;781401.
- Liskovykh M, Goncharov NV, Petrov N, Aksenova V, Pegoraro G, Ozbun LL, Reinhold WC, Varma S, Dasso M, Kumeiko V, Masumoto H, Earnshaw WC, Larionov V, Kouprina N. A novel assay to screen siRNA libraries identifies protein kinases required for chromosome transmission. Genome Res 2019;29(10):1719-1732.
- Zhang MS, Furuta M, Arnaoutov A, Dasso M. RCC1 regulates inner centromeric composition in a Ran-independent fashion. Cell Cycle 2018;17(6):739-748.
Collaborators
- Yoshiaki Azuma, PhD, University of Kansas, Lawrence, KS
- Beatriz Fontoura, PhD, University of Texas Southwestern Medical Center, Dallas, TX
- Natalay Kouprina, PhD, Developmental Therapeutics Branch, NCI, Bethesda, MD
- Vladimir Larionov, PhD, Developmental Therapeutics Branch, NCI, Bethesda, MD
- Brian C. Oliver, PhD, Laboratory of Cellular and Developmental Biology, NIDDK, Bethesda, MD
- Mihaela Serpe, PhD, Section on Cellular Communication, NICHD, Bethesda, MD
Contact
For more information, email mdasso@helix.nih.gov or visit http://sccr.nichd.nih.gov.


