Protein Sorting in the Endomembrane System

- Juan S. Bonifacino, PhD, Head, Section on Intracellular Protein Trafficking
- Rafael Mattera, PhD, Staff Scientist
- Xiaolin Zhu, Nurse, Technician
- Jing Pu, PhD, Research Fellow
- Raffaella De Pace, PhD, Visiting Fellow
- Saikat Ghosh, PhD, Visiting Fellow
- Carlos M. Guardia, PhD, Visiting Fellow
- Morie Ishida, PhD, Visiting Fellow
- Rui Jia, PhD, Visiting Fellow
- Tal Keren-Kaplan, PhD, Visiting Fellow
- Elodie Mailler, PhD, Visiting Fellow
- Amra Saric, PhD, Visiting Fellow
- Lucas M. Djavaherian, BA, Postbaccalaureate Student
- Gerard Walker, BA, Postbaccalaureate Student
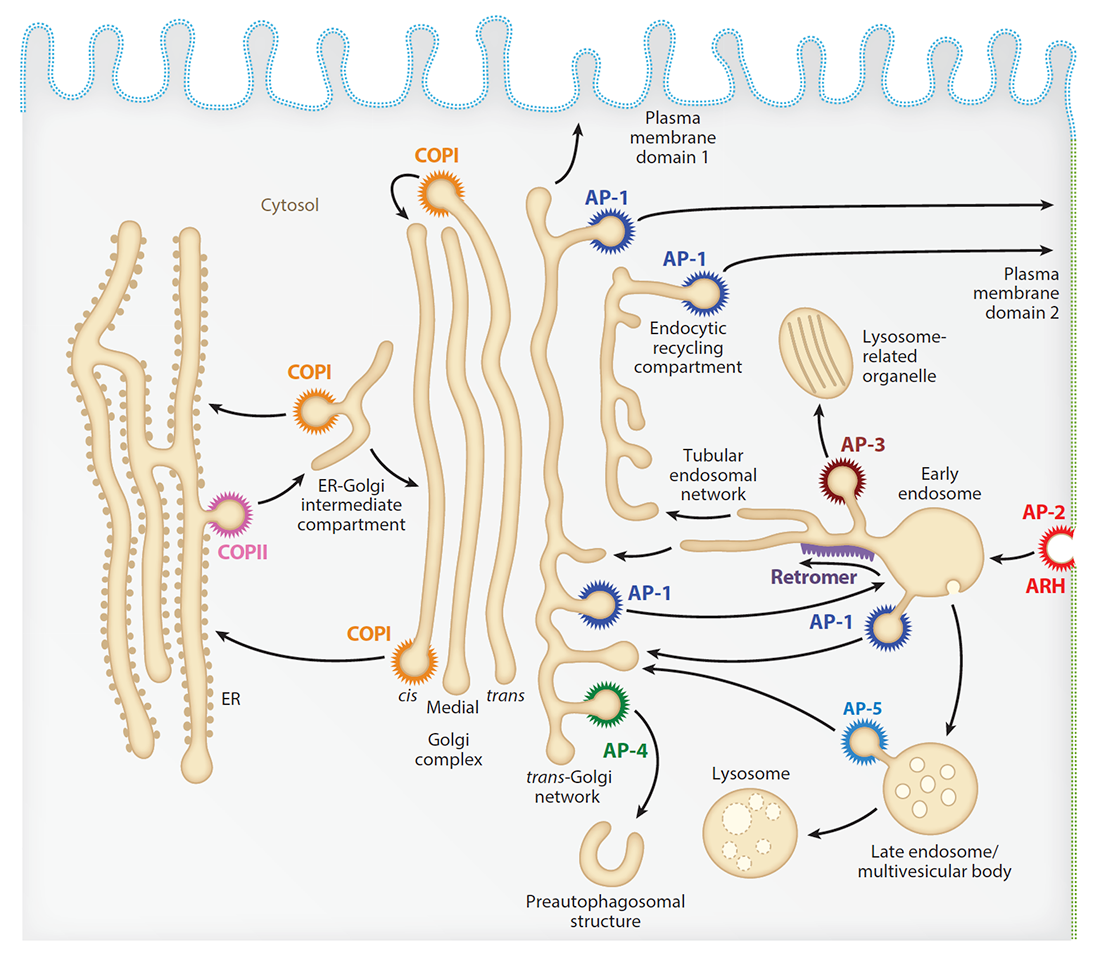
Our laboratory investigates the molecular mechanisms by which transmembrane proteins (referred to as “cargo”) are sorted to different compartments of the endomembrane system in eukaryotic cells. The system comprises an array of membrane-enclosed organelles including the endoplasmic reticulum (ER), the Golgi apparatus, the trans-Golgi network (TGN), endosomes, lysosomes, and lysosome-related organelles (LROs) (e.g., melanosomes), as well as different domains of the plasma membrane in polarized cells such as epithelial cells and neurons (Figure 1). Transport of cargo between the compartments is mediated by vesicular/tubular carriers that bud from a donor compartment, translocate through the cytoplasm, and eventually fuse with an acceptor compartment. Work in our laboratory focuses on the molecular machineries that mediate these processes, including (1) sorting signals and adaptor proteins that select cargo proteins for packaging into the transport carriers, (2) microtubule motors and organelle adaptors that drive movement of the transport carriers and other organelles through the cytoplasm, and (3) tethering factors that promote fusion of the transport carriers to acceptor compartments. We study the machineries in the context of different intracellular transport pathways, including endocytosis, recycling to the plasma membrane, retrograde transport from endosomes to the TGN, biogenesis of lysosomes and LROs, and polarized sorting in epithelial cells and neurons. Knowledge gained from this basic research is applied to the elucidation of disease mechanisms, including congenital disorders of protein trafficking, such as the pigmentation and bleeding disorder Hermansky-Pudlak syndrome (HPS), hereditary spastic paraplegias (HSPs), and ponto-cerebellar hypoplasias.
Click image to view.
Figure 1. Schematic representation of the endomembrane system of eukaryotic cells showing the localization of coats involved in protein sorting
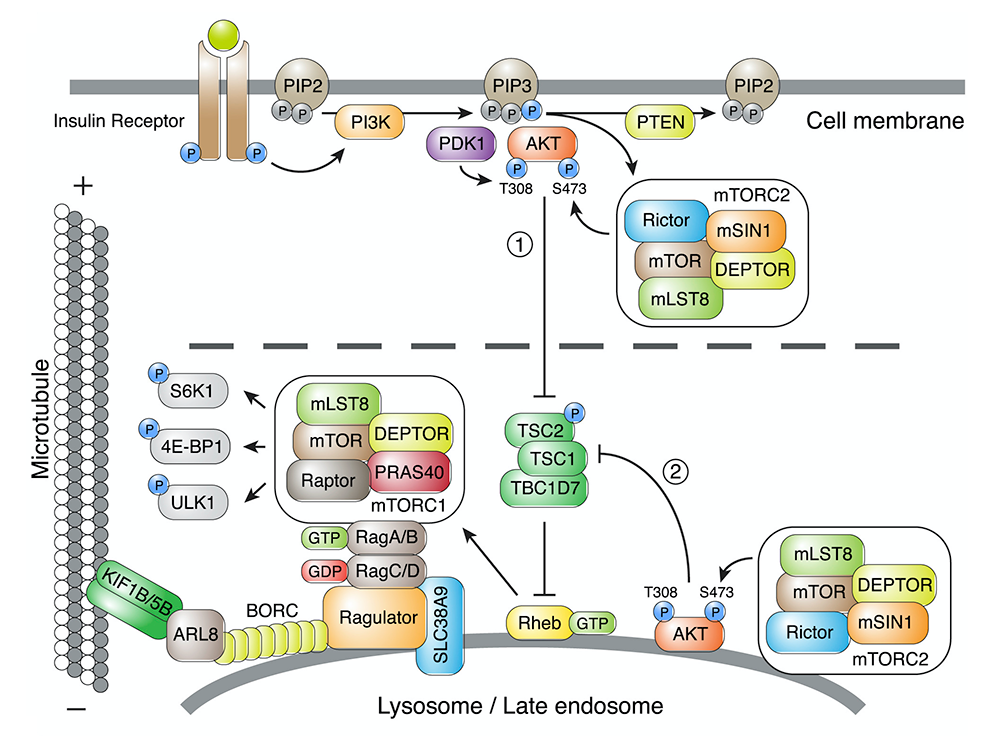
Lysosome positioning influences mTORC1 and mTORC2 signaling.
This past year, we examined the role of lysosome positioning on mTOR signaling in response to growth factors. Binding of growth factors to cognate receptors at the cell surface initiates intracellular signaling cascades that eventually reach organelles such as lysosomes. A key component of these signaling cascades is the serine/threonine kinase mTOR, which exists as a subunit of two complexes named mTORC1 and mTORC2 (Figure 2). Whereas mTORC1 associates with lysosomes, the intracellular distribution of mTORC2 is less well understood. We found that perinuclear clustering of lysosomes induced by uncoupling lysosomes from kinesin motors delayed the reactivation of mTORC1 caused by addition of serum (a source of growth factors). The finding indicated that increasing the distance of lysosome-associated mTORC1 from the source of growth-factor signaling at the plasma membrane delays the relay of signals through the cytoplasm. In addition, we made the surprising finding that mTORC2 reactivation after serum replenishment was also delayed by perinuclear clustering of lysosomes. The experiments demonstrated the existence of pools of both mTORC1 and mTORC2 that are sensitive to lysosome positioning, a finding that may explain how changes in lysosome positioning in cancer cells promote their proliferation.
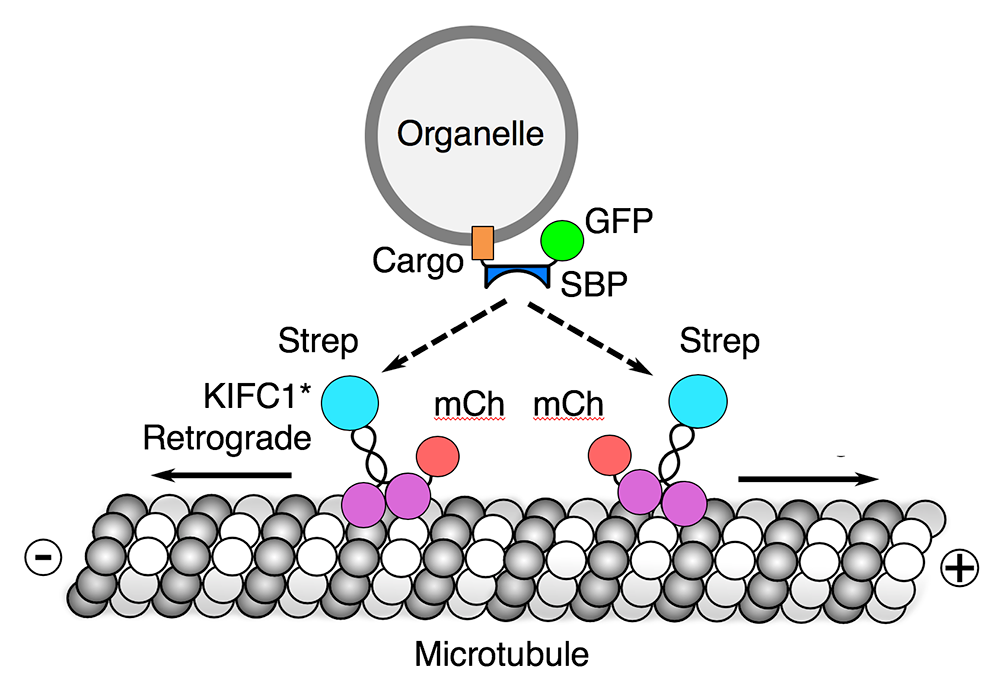
Reversible association with motor proteins (RAMP): a streptavidin-based method to manipulate organelle positioning
We also developed a novel method, named “reversible association with motor proteins” (RAMP), to manipulate organelle positioning within the cytoplasm (Figure 3). RAMP involves coexpressing (1) an organellar protein fused to the streptavidin-binding peptide (SBP), and (2) motor domains from plus-end–directed or minus-end–directed kinesin motors fused to streptavidin. The SBP–streptavidin interaction drives accumulation of organelles at the plus or minus end of microtubules, respectively. Importantly, addition of biotin dissociates the motor from the organelle, allowing restoration of normal patterns of organelle transport and distribution. We demonstrated that the method can be used to manipulate the distribution of various intracellular organelles, including lysosomes, mitochondria, peroxisomes, and the endoplasmic reticulum. The method should be useful for examining how the positioning of these organelles affects their functions, and for analyzing the movement of organelle cohorts upon release from the kinesin motor.
A novel neurodevelopmental disorder caused by mutations in the VPS51 subunit of the GARP and EARP complexes
The Golgi-associated retrograde protein (GARP) and endosome-associated recycling protein (EARP) complexes are related heterotetrameric complexes that associate with the TGN and recycling endosomes, respectively. GARP and EARP function to coordinate the SNARE–dependent fusion of endosome-derived transport carriers with their corresponding compartments, enabling retrograde transport to the TGN and recycling to the plasma membrane. GARP is composed of VPS51, VPS52, VPS53, and VPS54 subunits, whereas EARP is composed of VPS50, VPS51, VPS52, and VPS53 subunits. Although the complexes are known to play key roles in intracellular protein trafficking, their importance in human physiology remains poorly understood. In collaboration with David Everman, we recently identified compound heterozygous mutations in the gene encoding the shared GARP/EARP subunit VPS51 in a 6-year-old patient with severe global developmental delay, microcephaly, hypotonia, epilepsy, ponto-cerebellar abnormalities, liver dysfunction, lower extremity edema, and dysmorphic features. Biochemical and cellular analyses showed that the mutation in one allele causes a frameshift that produces a longer but highly unstable protein, whereas the mutation in the other allele produces a protein with a single amino-acid substitution that is stable but assembles less efficiently with the other GARP/EARP subunits. The mutations consequently result in reduced levels of GARP and EARP complexes in the patient’s cells. In addition, the patient's cells exhibit lysosomal abnormalities, consistent with the requirement of GARP for the sorting of acid hydrolases to lysosomes. The findings thus identified a novel genetic locus for a neurodevelopmental disorder and highlighted the critical importance of GARP/EARP function in cellular and organismal physiology.
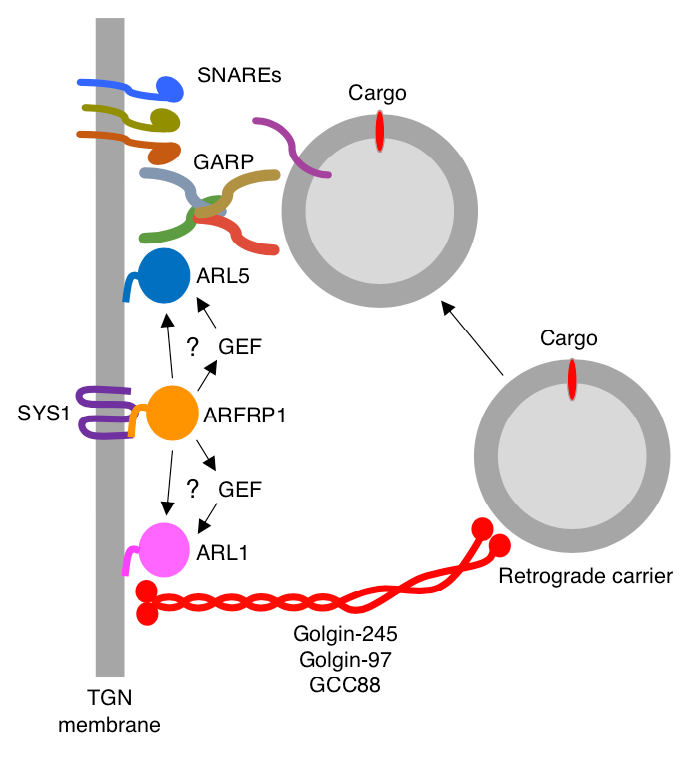
ARFRP1 functions upstream of both ARL1 and ARL5 to coordinate the recruitment of distinct classes of tethering factors to the TGN.
Despite the importance of GARP in cellular and organismal physiology, until recently it was unclear how it was recruited to the TGN and how its function was coordinated with that of a different class of tethering factors, long coiled-coil tethers of the golgin family. The golgins mediate long-distance capture of endosome-derived transport carriers, whereas GARP promotes SNARE–dependent fusion of the carriers with the TGN. This past year, we discovered that the ARF–like (ARL) GTPase ARFRP1 is an upstream activator of two other ARL GTPases, ARL1 and ARL5, which in turn recruit golgins and GARP, respectively, to the TGN (Figure 4). In addition, we found that this GTPase cascade is essential for the delivery of retrograde cargos to the TGN. From these findings, we concluded that ARFRP1 is a master regulator of retrograde-carrier tethering to the TGN. The mechanism involving the recruitment of distinct classes of tethering factors by a bifurcated GTPase cascade may be paradigmatic of other vesicular fusion events that take place within the cell.
Click image to view.
Figure 4. Model for the function of ARFRP1 in the coordinated recruitment of golgins and GARP to the TGN
The autophagy protein ATG9A promotes HIV-1 infectivity.
Nef is an accessory protein encoded by the primate immunodeficiency viruses HIV-1, HIV-2, and SIV and contributes to viral replication, assembly, budding, infectivity, and immune evasion, through engagement of various host-cell pathways. To gain a better understanding of the role of host-cell proteins in the functions of Nef, we carried out tandem affinity purification–mass spectrometry analysis and identified over 70 HIV-1 Nef–interacting proteins, including the autophagy-related 9A (ATG9A) protein. ATG9A is a transmembrane component of the machinery for autophagy, a catabolic process in which cytoplasmic components are degraded in lysosomal compartments. Pulldown experiments demonstrated that ATG9A interacts with Nef from not only HIV-1 and but also from SIV. However, expression of HIV-1 Nef had no effect on the levels and localization of ATG9A, or on autophagy, in the host cells. To investigate a possible role for ATG9A in virus replication, we used CRISPR/Cas9 to knock out (KO) ATG9A in HeLa cervical carcinoma and Jurkat T cells, and analyzed virus release and infectivity. We observed that ATG9A KO had no effect on the release of wild-type (WT) or Nef–defective HIV-1 in these cells. However, the infectivity of WT virus produced from ATG9A–KO HeLa and Jurkat cells was reduced 4-fold and 8-fold, respectively, compared with virus produced from WT cells. The reduction in infectivity was independent of the interaction of Nef with ATG9A and was not attributable to reduced incorporation of the viral envelope (Env) glycoprotein into the virus. The loss of HIV-1 infectivity was rescued by pseudotyping HIV-1 virions with the vesicular stomatitis virus G glycoprotein. From these studies, we concluded that ATG9A promotes HIV-1 infectivity in an Env–dependent but Nef–independent manner. ATG9A could promote infectivity by participating in either the removal of a factor that inhibits infectivity or in the incorporation of a factor that enhances infectivity of the viral particles. ATG9A is thus a novel host-cell factor implicated in HIV-1 infectivity.
Negative regulation of autophagy by UBA6-BIRC6–mediated ubiquitination of LC3
Although the process of autophagy has been extensively studied, the mechanisms that regulate it remain insufficiently understood. To identify novel autophagy regulators, we performed a whole-genome CRISPR/Cas9 knockout screen in H4 human neuroglioma cells expressing endogenous LC3B tagged with a tandem of GFP and mCherry. Using this methodology, we identified the ubiquitin-activating enzyme UBA6 and the hybrid ubiquitin-conjugating enzyme/ubiquitin ligase BIRC6 as autophagy regulators. We found that these enzymes cooperate to monoubiquitinate LC3B (a component of the autophagy machinery that participates in substrate selection and autophagosome formation), targeting it for proteasomal degradation (Figure 5). Knockout of UBA6 or BIRC6 increased autophagic flux under conditions of nutrient deprivation or protein synthesis inhibition. Moreover, depletion of UBA6 or BIRC6 reduced the formation of aggresome-like induced structures in H4 cells and α-synuclein aggregates in rat hippocampal neurons. The findings demonstrate that UBA6 and BIRC6 negatively regulate autophagy by limiting the availability of LC3B. Inhibition of UBA6/BIRC6 could be used to enhance autophagic clearance of protein aggregates in neurodegenerative disorders.
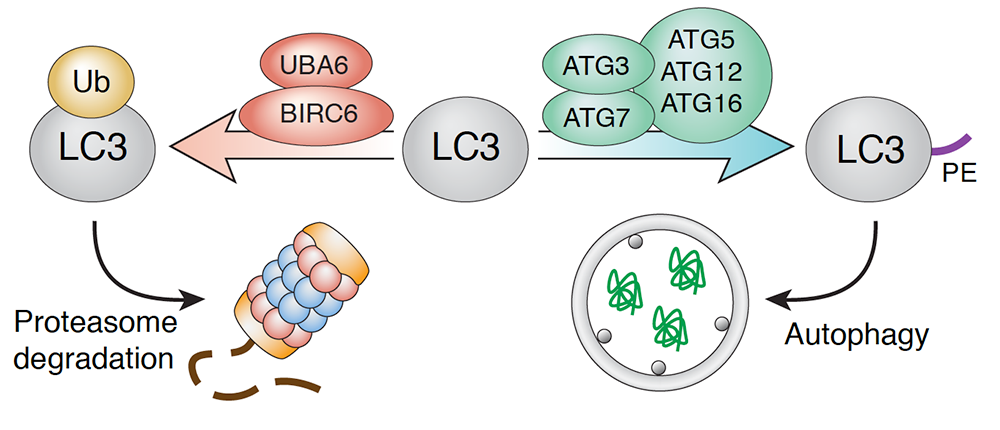
Click image to view.
Figure 5. Negative regulation of autophagy by UBA6-BIRC6–mediated ubiquitination of LC3
Publications
- Jia R, Bonifacino JS. Lysosome positioning influences mTORC2 and AKT signaling. Mol Cell 2019;75:26-38.
- Ishida M, Bonifacino JS. ARFRP1 functions upstream of ARL1 and ARL5 to coordinate recruitment of distinct tethering factors to the trans-Golgi network. J Cell Biol 2019;218(11):3681-3696.
- Guardia CM, De Pace R, Sen A, Saric A, Jarnik M, Kolin DA, Kunwar A, Bonifacino JS. Reversible association with motor proteins (RAMP): a streptavidin-based method to manipulate organelle positioning. PLoS Biol 2019;17:e3000279.
- Gershlick DC, Ishida M, Jones JR, Bellomo A, Bonifacino JS, Everman DB. A neurodevelopmental disorder caused by mutations in the VPS51 subunit of the GARP and EARP complexes. Hum Mol Genet 2019;28:1548-1560.
- Dell'Angelica EC, Bonifacino JS. Coatopathies: genetic disorders of protein coats. Annu Rev Cell Dev Biol 2019;35:131-168.
Collaborators
- Melvin DePamphilis, PhD, Section on Eukaryotic DNA Replication, NICHD, Bethesda, MD
- David Everman, MD, Greenwood Genetic Center, Greenwood, SC
- Eric Freed, PhD, Retroviral Replication Laboratory, Center for Cancer Research, NCI, Frederick, MD
- Sergio Grinstein, PhD, Hospital for Sick Children, Toronto, Canada
Contact
For more information, email bonifacinoj@helix.nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/bonifacino.