Chromatin Remodeling and Gene Activation

- David J. Clark, PhD, Head, Section on Chromatin and Gene Expression
- Peter Eriksson, PhD, Staff Scientist
- Razvan V. Chereji, PhD, Research Fellow
- Josefina Ocampo, PhD, Visiting Fellow
- Sean C. Clark, MS, Postbaccalaureate Fellow
- Allison F. Dennis, BS, Graduate Student
Aberrant gene regulation is the basis of many disease states. Our main objective is to understand how genes are activated for transcription in the context of chromatin structure. Chromatin is not just a packaging system for DNA in eukaryotic cells—it also participates in gene regulation. The structural subunit of chromatin is the nucleosome, which contains nearly two turns of DNA coiled around a central core histone octamer. Nucleosomes are generally quite regularly spaced along the DNA, like beads on a string. Gene regulation involves either attenuation of the inherently repressive properties of nucleosomes to facilitate gene expression, or enhancement of those properties to ensure complete repression. These events are choreographed by DNA sequence–specific transcription factors (activators and repressors) and chromatin-remodeling complexes. The latter can be divided into two groups: histone or DNA–modifying enzymes that implement the “epigenetic code,” and ATP–dependent remodeling machines, which move or displace nucleosomes. Genes encoding subunits of some of these enzymes are often mutated in various cancers.
We are exploiting high-throughput technologies to obtain genome-wide maps of nucleosomes, chromatin-remodeling complexes and RNA polymerase II in budding yeast to determine what happens to nucleosomes on genes when these are activated. We find that transcription results in disruption and loss of some nucleosomes on the gene with re-positioning of the remaining nucleosomes (Figure 1). The current objectives of our yeast studies are to: (1) determine the roles of various chromatin-remodeling complexes (RSC, SWI/SNF, ISW1, ISW2, and CHD1) in chromatin organization and gene expression; (2) understand the dynamics of transcription by RNA polymerase II through chromatin in vivo, with an emphasis on potential regulation of the dissociation of polymerase after transcription.
Most of our work involves the use of Saccharomyces cerevisiae (budding yeast) as a model organism, but, through collaborations, we are now extending our studies of chromatin remodeling from yeast to mouse. Mammalian systems are much more complex and challenging than yeast. We are investigating the effects of dexamethasone on mouse-cell chromatin in a collaboration with the Hager lab. We are also comparing the chromatin structures of neurons, oligodendrocytes, and astrocytes in a collaboration with the Fields lab.
Click image to enlarge.
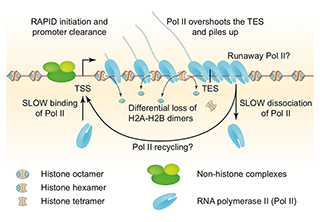
Figure 1. Chromatin structure of a heavily transcribed gene
The nucleosome-depleted region (NDR) at the promoter is expanded upstream to accommodate additional non-histone complexes (both H4 and H2B are partially lost). RNA polymerase II (Pol II) binds near the transcription start site (TSS) and clears the promoter rapidly (low Pol II at the TSS). Pol II levels over the remainder of the gene are high and coincide with some loss of H4 and heavy loss of H2B, resulting in sub-nucleosomal particles (tetrasomes and hexasomes). Pol II overshoots the transcript end site (TES) and accumulates about 140 bp downstream, sometimes farther (“runaway” Pol II). Accumulation of Pol II downstream of the TES implies that dissociation from DNA is relatively slow, such that more Pol II queues up behind it. Slow dissociation might facilitate transfer of Pol II back to the promoter (“recycling”), increasing the transcription rate. Adapted from Cole HA et al. Nucl Acids Res 2014;42:12512.
The ISW1 and CHD1 ATP–dependent chromatin remodelers compete to set nucleosome spacing in vivo.
ATP–dependent chromatin-remodeling machines play a central role in gene regulation by manipulating chromatin structure. Most genes have a nucleosome-depleted region at the promoter and an array of regularly spaced nucleosomes phased relative to the transcription start site. In vitro, the three known yeast nucleosome-spacing enzymes (CHD1, ISW1, and ISW2) form arrays with different spacing. In this study, we used genome-wide nucleosome sequencing (MNase-seq) to determine whether these enzymes space nucleosomes differently in vivo. Specifically, we constructed all combinations of isw1, isw2, and chd1 null mutants and compared their chromatin structures. We found that CHD1 and ISW1 compete to set the spacing on most genes, such that CHD1 dominates genes with shorter spacing and ISW1 dominates genes with longer spacing. In contrast, ISW2 plays a minor role, limited to transcriptionally inactive genes. Heavily transcribed genes show weak phasing and extreme spacing, either very short or very long, and are depleted of linker histone (H1). Genes with longer spacing are enriched in H1, which directs chromatin folding. We propose that CHD1 directs short spacing, resulting in eviction of H1 and chromatin unfolding, whereas ISW1 directs longer spacing, allowing H1 to bind and condense the chromatin (Reference 1). Thus, competition between the two remodelers to set the spacing on each gene may result in a highly dynamic chromatin structure (Figure 2). We are currently testing this model.
Click image to enlarge.
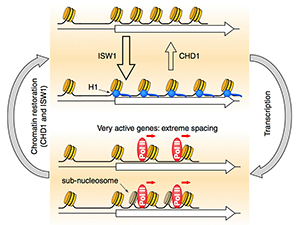
Figure 2. Roles of the CHD1, ISW1, and ISW2 nucleosome spacing enzymes in creating a dynamic chromatin structure
We propose that CHD1 and ISW1 compete to determine the spacing on most genes. Genes dominated by ISW1 have longer spacing, resulting in linkers long enough to bind to H1 (blue circles) with high affinity and thus in more condensed chromatin. Genes dominated by CHD1 have short spacing, with linkers too short for H1 binding. Competition between CHD1 and ISW1 occurs on most genes, resulting in intermediate spacing, possibly creating a highly dynamic chromatin structure. The most inactive genes are also affected by ISW2, resulting in slightly longer spacing. Heavily transcribed genes have highly disrupted chromatin, including gaps created by loss of some nucleosomes and sub-nucleosomes (darker cylinders) and resulting from displacement of H2A-H2B dimers and thus in extreme spacing.
Genome-wide cooperation by HAT Gcn5, remodeler SWI/SNF, and chaperone Ydj1 in promoter nucleosome eviction and transcriptional activation
Gene activation generally involves nucleosome removal and/or nucleosome shifts in the promoter region. Histone chaperones, ATP–dependent nucleosome-remodeling complexes and histone acetyltransferases (HATs) have been implicated in nucleosome disassembly at promoters of particular yeast genes. However, it is unclear whether these co-factors function ubiquitously. Furthermore, the impact of nucleosome eviction at promoters on transcription genome-wide is poorly understood. In a collaboration with the Hinnebusch lab, we used chromatin immunoprecipitation (ChIP-seq) of histone H3 and RNA polymerase II (Pol II) in mutants lacking single or multiple co-factors to address these issues for about 200 genes belonging to the Gcn4 transcriptome. We find that about 70 genes exhibit marked reductions in H3 promoter occupancy on induction by amino-acid starvation. A close examination of four target genes in a panel of mutants indicated that the SWI/SNF ATP–dependent chromatin remodeler, the Gcn5 HAT, a subunit of the SAGA complex, the heat-shock protein Hsp70 co-chaperone Ydj1, and the chromatin-associated factor Yta7 are required downstream of Gcn4 binding, whereas other co-factors (the Asf1 and Nap1 histone chaperones, the Rtt109 HAT, the RSC chromatin remodeler, and the H2AZ histone variant) are dispensable for robust H3 eviction in otherwise wild-type cells. Using ChIP-seq to interrogate all 70 exemplar genes in single, double, and triple mutants, we found that Gcn5, Snf2, and Ydj1 were implicated in H3 eviction at most, but not all Gcn4 target promoters, with Gcn5 generally playing the greatest role and Ydj1 the least. Remarkably, the three co-factors cooperate similarly in H3 eviction at virtually all yeast promoters. Defective H3 eviction in co-factor mutants was coupled with reduced Pol II occupancies for the Gcn4 transcriptome and the most highly expressed un-induced genes, but the relative Pol II levels at most genes were unaffected or even elevated. The findings indicate that nucleosome eviction is crucial for robust transcription of highly expressed genes, but that other steps in gene activation are more rate-limiting for most other yeast genes (Reference 2).
MNase–sensitive complexes in yeast: nucleosomes and non-histone barriers
Micrococcal nuclease (MNase) is commonly used to map nucleosomes genome-wide, but nucleosome maps are affected by the degree of digestion. It has been proposed that many yeast promoters are not nucleosome-free but occupied by easily digested, unstable, “fragile” nucleosomes. We analyzed the histone content of all MNase–sensitive complexes by MNase–ChIP-seq and Sonication–ChIP-seq. We found that yeast promoters are predominantly bound by non-histone protein complexes, with little evidence for binding by fragile nucleosomes. We did detect MNase–sensitive nucleosomes elsewhere in the genome, including at transcription termination sites. However, they have high A/T content, suggesting that MNase sensitivity does not indicate instability but a preference of MNase for A/T–rich DNA, such that A/T–rich nucleosomes are digested faster than G/C–rich nucleosomes. We confirmed our observations by analyzing ChIP-exo, chemical mapping, and ATAC–seq data from other laboratories. Thus, histone ChIP-seq experiments are essential to distinguish nucleosomes from other DNA–binding proteins that protect against MNase. A manuscript describing this work has been submitted (Chereji RV, Ocampo J, Clark DJ).
Chromatin remodeling in neurons and oligodendroglia
The aim of this project, a collaboration with the Fields lab, is to determine the relationship between chromatin organization and gene expression in dorsal root ganglion neurons and oligodendroglial cells. Neuronal chromatin is remarkably atypical: the linker DNA between nucleosomes is much shorter than in other cell types, including oligodendroglial cells, and neurons are deficient in histone H1. The global organization of neuronal chromatin is therefore very different from other cells, but the significance of such organization is unknown. We are testing the relationship between global chromatin organization and transcription. We have mapped nucleosomes genome-wide in isolated mouse neurons and in oligodendroglial cells, using paired-end sequencing, and measured gene expression by RNAseq. We are now correlating their chromatin structures with transcriptional activity. Our experiments should provide insight into why neuronal chromatin organization is so different from that of other cells, including glia.
Publications
- Ocampo J, Chereji RV, Eriksson PR, Clark DJ. The ISW1 and CHD1 ATP-dependent chromatin remodelers compete to set nucleosome spacing in vivo. Nucl Acids Res 2016;44:4625-4635.
- Qiu H, Chereji RV, Hu C, Cole HA, Rawal Y, Clark DJ, Hinnebusch AG. Genome-wide cooperation by HAT Gcn5, remodeler SWI/SNF and chaperone Ydj1 in promoter nucleosome eviction and transcriptional activation. Genome Res 2016;26:211-225.
- Ocampo J, Clark DJ. A positive twist to the centromeric nucleosome. Cell Rep 2015;13:645-646.
- Gerasimova NS, Pestov NA, Kulaeva OI, Clark DJ, Studistky VM. Transcription-induced DNA supercoiling: new roles of intranucleosomal DNA loops in DNA repair and transcription. Transcription 2016;7:91-95.
- Ocampo J, Cui F, Zhurkin VB, Clark DJ. The proto-chromatosome—a fundamental subunit of chromatin? Nucleus 2016;7:382-387.
Collaborators
- Feng Cui, PhD, Rochester Institute of Technology, Rochester, NY
- Douglas Fields, PhD, Section on Nervous System Development and Plasticity, NICHD, Bethesda, MD
- Gordon L. Hager, PhD, Laboratory of Receptor Biology and Gene Expression, Center for Cancer Research, NCI, Bethesda, MD
- Alan G. Hinnebusch, PhD, Section on Nutrient Control of Gene Expression, NICHD, Bethesda, MD
- James Iben, PhD, Section on Molecular Dysmorphology, NICHD, Bethesda, MD
- Philip Lee, PhD, Section on Nervous System Development and Plasticity, NICHD, Bethesda, MD
- Vasily M. Studitsky, PhD, Fox Chase Cancer Center, Temple University Health System, Philadelphia, PA
- Victor Zhurkin, PhD, Laboratory of Cell Biology, Center for Cancer Research, NCI, Bethesda, MD
Contact
For more information, email clarkda@mail.nih.gov or visit http://clarklab.nichd.nih.gov.


