Interplay between Membrane Organelles, Cytoskeleton, and Metabolism in Cell Organization and Function

- Jennifer Lippincott-Schwartz, PhD, Head, Section on Organelle Biology
- Sarah Cohen, PhD, Visiting Fellow
- Carolyn Ott, PhD, Staff Scientist
- Christopher Obara, PhD, Postdoctoral Fellow
- Timothy Petri, PhD, Postdoctoral Fellow
- Prabuddha Sengupta, PhD, Postdoctoral Fellow
- Arnold Seo, PhD, Postdoctoral Fellow
- Alex Valm, PhD, Postdoctoral Fellow
- Aubrey Weigel, PhD, Postdoctoral Fellow
- Lingfeng Chen, PhD, Volunteer
- Prasanna Satpute, PhD, Volunteer
- Alex Ritter, BA, Graduate Student
- Bennett Waxse, BA, Graduate Student
We investigate the global principles underlying cell behavior at both small and large spatial scales. At the small scale, we employ the super-resolution imaging techniques of photoactivated localization microscopy (PALM), interferometric 3D PALM, single-particle tracking PALM, and pair-correlation PALM to map the spatial organization, stoichiometry, and dynamics of proteins associated with various membrane-bound compartments and with the cytoskeleton. We also employ fluorescence photobleaching, photoactivation, fluorescence correlation, and fluorescence energy transfer methods to measure protein-protein interactions, protein turnover rates, and protein association rates. Such approaches allow us to assay cellular functions, including receptor stoichiometry and protein clustering and diffusion behavior at the nanometric scale in living cells. At the large scale, we investigate how complex behaviors of cells arise, such as cell crawling, polarization, cytokinesis, and viral budding. We study these complex behaviors by quantitatively analyzing diverse intracellular processes, including membrane trafficking, autophagy, actin/microtubule dynamics, and organelle assembly/disassembly pathways, which undergo dramatic changes as cells alter their behavior and organization throughout life. To assist these efforts, we combine various fluorescence-based imaging approaches, including total internal reflection fluorescence (TIRF) microscopy imaging and spinning-disk and laser-scanning confocal microscopy, with FRAP (fluorescence recovery after photobleaching), FLIP (fluorescence loss in photobleaching), and photoactivation to obtain large image data sets. We process the data sets computationally to extract biochemical and biophysical parameters, which can be related to the results of conventional biochemical assays. We then use the results to generate mechanistic understanding and predictive models of the behavior of cells and subcellular structures (including endoplasmic reticulum, Golgi, cilia, endosomes, lysosomes, autophagosomes, and mitochondria) under healthy and pathological conditions.
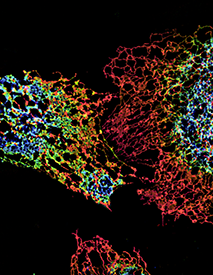
Click image to enlarge.
ER structure visualized using structured illumination microscopy (SIM)
Structure of peripheral ER in three cells labeled with an ER protein marker visualized by three-dimensional SIM. The color coding represents different z heights of the ER.
ER structure and dynamics visualized with increased spatio-temporal resolution
The endoplasmic reticulum (ER) consists of interconnected tubules and flattened sheets that extend from the nuclear envelope to the periphery of the cell, impacting every cellular compartment through its contacts and functional interactions. Mutations in proteins that regulate the shape of the ER lead to various neurological disorders. We employed five different super-resolution technologies with complementary strengths and weaknesses in spatial and temporal capabilities to study the fine morphology and dynamics of the peripheral ER. A high-speed variation of structured illumination microscopy (SIM) allowed ER dynamics to be visualized at unprecedented speeds and resolution. Three-dimensional SIM (3D-SIM) and Airyscan imaging allowed comparison of the fine distributions of different ER–shaping proteins. Lattice light sheet point accumulation for imaging in nanoscale topography (LLS-PAINT) and focused ion-beam scanning electron microscopy (FIB-SEM) permitted 3D characterization of various ER structures. Using these approaches, we observed that many ER structures previously proposed to be flat membrane sheets are instead densely packed tubular arrays, which we call ER matrices. The matrices were extremely compact, with spaces between tubules below the resolving power of most super-resolution methodologies. We also discovered that ER tubules and junctions undergo rapid oscillations, rapidly interconverting from tight to loose arrays. The oscillations of tubules and junctions were energy dependent and allowed the ER to interconvert between tight and loose tubule networks. Our finding of dense tubular matrices in areas previously thought of as flat sheets provides a new model for maintaining and generating ER structure. In this model, ER matrices would sequester excess membrane proteins and lipids, and their dynamic interconversion into loose tubule arrays would permit the ER to rapidly extend its shape to reach the cell periphery, for example during cell locomotion.
ER trapping reveals that Golgi enzymes continually revisit the ER through a recycling pathway that controls Golgi organization.
The Golgi apparatus is the major processing and sorting station at the crossroads of the secretory pathway, containing specialized sorting and transport machinery, which either drive secretory trafficking or filter out selected membrane and protein components for return to the ER for continued use. How the Golgi maintains its structure and function amidst this ongoing bi-directional membrane trafficking has been the subject of a long-standing debate. In particular, it is unclear whether Golgi enzymes remain localized within the Golgi or constitutively cycle through the ER.
To address this question, we used a rapamycin trapping assay to test whether Golgi enzymes become trapped in the ER upon expression of both an ER–trapping protein and Golgi enzyme bait. We found that within four hours of rapamycin treatment, Ii-FKBP12 (i.e., ER trapper) trapped nearly all Golgi-localized FRB–Golgi enzyme (i.e., Golgi bait) in the ER. Direct redistribution from the Golgi to the ER during rapamycin treatment occurred because selective photoactivation of FRB–tagged, photoactivatable Golgi enzyme in the Golgi resulted in the signal shifting to the ER. In the ER, fluorescent forms of FRB–Golgi enzyme and Ii-FKBP12 underwent FRET, indicating direct binding upon rapamycin-induced redistribution. By contrast, use of Ii-FRB, which we showed is an inefficient ER trap, resulted in minimal redistribution of FKBP12–tagged Golgi enzymes during rapamycin treatment. These data demonstrated that Golgi enzymes constitutively cycle through the ER. Using our trapping scheme, we further identified roles of Rab6a and iPLA2 in Golgi enzyme recycling and showed that retrograde transport of Golgi membrane is necessary for Golgi dispersal during microtubule depolymerization and mitosis.
Publications
- Sengupta P, Satpute-Krishnan P, Seo AY, Burnette DT, Patterson GH and Lippincott-Schwartz J. ER trapping reveals Golgi enzymes continually revisit the ER through a recycling pathway that controls Golgi organization. Proc Natl Acad Sci USA 2015;112:6752-6761.
- Yao PJ, Petralia RS, Ott C, Wang YX, Lippincott-Schwartz J, Mattson MP. Dendrosomatic Sonic Hedgehog signaling in hippocampal neurons regulates axon elongation. J Neurosci 2015;35:16126-16141.
- Rikhy R, Mavrakis M, Lippincott-Schwartz J. Dynamin regulates metaphase furrow formation and plasma membrane compartmentalization in the syncytial Drosophila embryo. Biol Open 2015;4:301-311.
- Nixon-Abell J, Obara CJ, Weigel AV, Li D, Legant WR, Xu CS, Pasolli HA, Harvey K, Hess HF, Betzig E, Blackstone C, Lippincott-Schwartz J. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science 2016;354(6311):Epub ahead of print.
- Sulkowski MJ, Han TH, Ott C, Wang Q, Verheyen EM, Lippincott-Schwartz J, Serpe M. A novel, noncanonical BMP pathway modulates synapse maturation at the Drosophila neuromuscular junction. PLoS Genet 2016;12(1):e1005810.
Collaborators
- Eric Betzig, PhD, Howard Hughes Medical Institute, Janelia Farm Research Campus, Ashburn, VA
- Craig Blackstone, MD, PhD, Cell Biology Section, NINDS, Bethesda, MD
- Yu-Tsung Chen, PhD, Section on Cell Cycle Regulation, NICHD, Bethesda, MD
- Harald Hess, PhD, Howard Hughes Medical Institute, Janelia Farm Research Campus, Ashburn, VA
- Mark P. Matteson, PhD, Laboratory of Neurosciences, NIA, Baltimore, MD
- Jonathon J. Nixon-Abell, PhD, Cell Biology Section, NINDS, Bethesda, MD
- George Patterson, PhD, Section on Biophotonics, NIBIB, Bethesda, MD
- Richa Rikhy, PhD, Indian Institute of Science Education and Research (IISER), Pune, India
- Mihaela Serpe, PhD, Unit on Cellular Communication, NICHD, Bethesda, MD
- Pamela J. Yao, PhD, Cellular and Molecular Neurosciences Section, NIA, Baltimore, MD
Contact
For more information, email lippincj@mail.nih.gov or visit http://lippincottschwartzlab.nichd.nih.gov.


