Host-pathogen interactions during Legionella pneumophila infection

- Matthias Machner, PhD, Head, Unit on Microbial Pathogenesis
- Eric Cheng, PhD, Visiting Fellow
- Nicole Ellis, PhD, Visiting Fellow
- Pei-Chung Lee, PhD, Visiting Fellow
- Yi-han Lin, PhD, Visiting Fellow
- Xiao Li, PhD, Visiting Fellow
- Katherine Gillis, BA, Postbaccalaureate Student
Our main research goal is to obtain mechanistic insight into the virulence strategies of microbial pathogens. As a model organism we use the bacterium Legionella pneumophila, the causative agent of a potentially fatal respiratory infection known as Legionnaires' disease. Contrary to what its name may imply, Legionnaires’ disease occurs in individuals of all ages, including children who receive respiratory therapy, newborns who recently underwent surgery or under-water birth, and children who are immune-compromised. We are committed to an in-depth analysis of mechanisms that allow L. pneumophila to exploit the human host and cause disease. Insights gained from these studies will ultimately improve our ability to better diagnose, prevent, and fight Legionnaires’ disease and related illnesses, thereby contributing to the success of NICHD’s mission.
Upon inhalation of contaminated water droplets, L. pneumophila enters the lung and is phagocytosed (taken up) by alveolar macrophages, specialized immune cells. Instead of being degraded by these cells, the pathogen establishes a protective membrane compartment, the Legionella-containing vacuole (LCV). Within this intravacuolar niche, L. pneumophila can replicate to high numbers before killing the host cell and infecting neighboring cells.
Intracellular survival of L. pneumophila depends on the activity of more than 300 proteins, or effectors, that are injected into the host cell, where they create conditions favorable for infection. L. pneumophila mutants that are defective in effector protein delivery fail to escape endo-lysosomal degradation, underscoring the key role of microbial effectors for bacterial virulence. Our aim is to obtain a detailed mechanistic insight into the regulation and function of L. pneumophila effectors by investigating host-pathogen interactions at a molecular, cellular, and structural level. Deciphering the virulence program of this dangerous pathogen will set the stage for the development of novel therapeutics aimed at treating or preventing Legionnaires' disease and related illnesses.
Host-pathogen interaction profiling using self-assembling human protein arrays
Click image to enlarge.
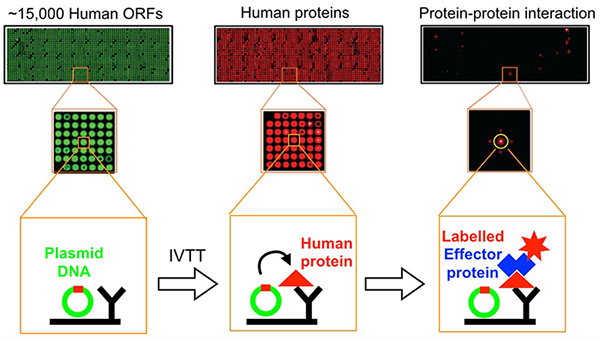
Figure 1. Flow scheme of NAPPA fabrication and protein interaction assay
NAPPA arrays are converted from DNA to protein through in vitro transcription/translation. Newly synthesized tagged human proteins are captured on the array by a tag-specific antibody, and binding of HaloTag-query protein to its target on NAPPA is detected using Alexa660–labeled Halo-ligand. Plasmid cDNA on the array is stained green, proteins red.
Despite many years of intense study by several groups, L. pneumophila effectors of unknown function vastly outnumber those that have been well characterized. The identification of host targets has remained particularly challenging owing to the lack of simple detection tools that avoid abundance biases while providing an open format for experimental modifications. Together with the group of Joshua LaBaer, we established an improved protein-protein interaction platform called Nucleic Acid–Programmable Protein Array (NAPPA). For the array, thousands of genes encoding tagged human bait proteins are printed on an aminosilane-coated slide (Figure 2). At the time of assay, the proteins are freshly synthesized through in vitro transcription/translation (IVTT) and displayed in situ using co-spotted anti-tag antibodies.
We developed an improved NAPPA by introducing the HaloTag (Promega) at the C-terminus of the bacterial query protein. HaloTag is a modified haloalkane dehalogenase designed to covalently bind to synthetic Halo-ligands (haloalkanes). Once applied to NAPPA, binding of a HaloTag-query protein to its interactor(s) can be specifically detected among thousands of proteins using an Alexa660–labeled Halo-ligand. In a proof-of-concept study, we probed the NAPPA with the L. pneumophila effectors SidM or LidA and identified most of the known targets but also potential novel interaction candidates, a subset of which we confirmed in independent in vitro pull-down and in vivo cell-based assays. The NAPPA approach also facilitated the discovery of two novel interaction partners for AnkX, a Legionella effector protein involved in lysosomal avoidance during intracellular replication. We confirmed these interactions through both independent in vitro pull-down and cell-based assays. Protein truncation studies revealed that association with both host targets is mediated by the N-terminal region of AnkX, while the C-terminal region tethers AnkX to membranes. Thus, the NAPPA technology represents a platform for the easy and reliable discovery of host targets for effector proteins from L. pneumophila, and can be easily adapted to the study of proteins from other microbial pathogens.
Click image to enlarge.
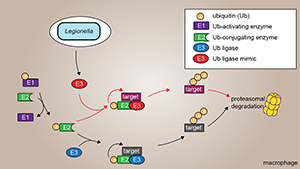
Figure 2. Hijacking of host ubiquitination by Legionella E3 ligase mimics
Ubiquitination of target proteins is mediated by the sequential action of three types of enzyme: Ub–activating enzymes (E1s), Ub–conjugating enzymes (E2s), and Ub ligases (E3s). The E1 activates Ub in an ATP–dependent manner and transfers Ub onto an E2. From there, Ub is transferred onto a target protein by an E3 ligase, often resulting in proteasomal degradation of the target. Legionella exploits the host ubiquitination machinery by producing its own E3 ligase mimics, which hijack the E2–Ub complex to target different host proteins for ubiquitination and subsequent degradation.
Strategy of molecular mimicry to hijack host-cell ubiquitination
Bacterial pathogens often target conserved host pathways by encoding proteins that are molecular mimics of cellular enzymes, thus tricking the host cell into surrendering its resources to the bacteria. We discovered that L. pneumophila uses such a strategy to exploit ubiquitination, a conserved post-translational modification that is mediated by a family of enzymes called E3 ubiquitin ligases. L. pneumophila encodes its own molecular mimics of E3 ligases, including the effector protein RavN, thereby subverting the ubiquitin pathway for its own benefit during infection. By testing truncated RavN variants in an in vitro reconstitution assay, we found that the E3 ligase activity of RavN is located within its N-terminal region. Using protein crystallography, we revealed that the fold of RavN shows only residual resemblance to conventional eukaryotic E3s. The N-terminal region of RavN displays a U-box-like motif that lacks the central alpha helix commonly found in other U-box domains, indicating that RavN is an E3 ligase relic that has undergone significant evolutionary alteration. Yet its mode of interaction with E2 enzymes, host proteins that are important for the ubiquitin transfer reaction, has been preserved throughout evolution, and substitution of amino acid residues within the predicted E2 binding interface have rendered RavN inactive.
Using RavN as a model for an in silico analysis, we discovered several additional E3 ligase mimics within the effector repertoire of L. pneumophila that, similar to RavN, lack significant homology to known E3s but, nonetheless, catalyze the ubiquitination reaction. Our findings support the hypothesis that E3 ligases have been a vital part of the virulence program of L. pneumophila and that these effectors, despite having undergone extensive evolutionary changes, have retained features that are critical for their biological function, including the ability to hijack factors that are part of the host ubiquitination machinery. These data indicate that ubiquitination is more extensively exploited during infection by L. pneumophila than anticipated and that interference with this post-translational modification could be a novel therapeutic strategy to antagonize infections by L. pneumophila and related pathogens.
Publications
- Lin YH, Doms AG, Cheng E, Kim B, Evans TR, Machner MP. Host cell-catalyzed S-palmitoylation mediates Golgi targeting of the Legionella ubiquitin ligase GobX. J Biol Chem 2015; 290:25766-25781.
- Yu X, Decker KB, Baker K, Neunuebel MR, Graves M, Westcott N, Hang H, LaBaer J, Qiu J, Machner MP. Host-pathogen interaction profiling using self-assembling human protein arrays. J Proteome Res 2015; 14:1920-1936.
- Machner MP, Storz G. Infection biology: small RNA with a large impact (invited commentary). Nature 2016;529:472-473.
Collaborators
- Aitor Hierro, PhD, CIC bioGUNE Institute, Bilbao, Spain
- Joshua LaBaer, MD, PhD, Virginia G. Piper Center for Personalized Diagnostics, Arizona State University, Tempe, AZ
Contact
For more information, email machnerm@mail.nih.gov or visit http://machnerlab.nichd.nih.gov.


