Chromatin Remodeling and Gene Activation

- David J. Clark, PhD, Head, Section on Chromatin and Gene Expression
- Peter Eriksson, PhD, Staff Scientist
- Christopher Coey, PhD, Intramural Research Training Award Fellow
- Hemant K. Prajapati, PhD, Visiting Fellow
- Zhuwei Xu, PhD, Visiting Fellow
- Paul Elizalde, BS, Postbaccalaureate Fellow
Aberrant gene regulation is the basis of many disease states. Our main objective is to understand how genes are activated for transcription in the context of chromatin structure. Chromatin is not solely a packaging system for DNA in eukaryotic cells, but also participates in gene regulation. The structural subunit of chromatin is the nucleosome, which contains nearly two turns of DNA coiled around a central core histone octamer. Nucleosomes are generally quite regularly spaced along the DNA, like beads on a string. Gene regulation involves either attenuation of the inherently repressive properties of nucleosomes to facilitate gene expression, or enhancement of those properties to ensure complete repression. These events are choreographed by DNA sequence–specific transcription factors (activators and repressors) and chromatin-remodeling complexes. The latter can be divided into two groups: histone- or DNA–modifying enzymes that implement the “epigenetic code”, and ATP–dependent remodeling machines that move or displace nucleosomes. We are exploiting and developing high-throughput technologies to obtain genome-wide maps of nucleosomes, chromatin-remodeling complexes, and RNA polymerase II in budding yeast to determine how chromatin organization is affected when genes are activated. The current objectives of our yeast studies are to: (1) determine the roles of the various chromatin-remodeling complexes (RSC, SWI/SNF, ISW1, ISW2, CHD1, Ino80C) in chromatin organization and gene expression; why there are so many different remodelers; whether they are functionally redundant, our studies so far indicating that each remodeling enzyme makes a different contribution to chromatin organization; (2) test the hypothesis that nucleosomes control DNA accessibility and play a vital role in gene regulation by blocking promoters (Figure 1). We are extending our studies of chromatin remodeling from yeast to higher organisms.
Many human diseases have been linked to chromatin-remodeling enzymes and epigenetic modifications. For example, mutations in the gene encoding the hSNF5 subunit of the SWI/SNF ATP–dependent chromatin-remodeling complex are strongly linked to pediatric rhabdoid tumors. The CHD (chromatin helicase DNA binding) class of ATP–dependent remodelers has also been linked to cancer and autism. Cancer therapies and drugs aimed at epigenetic targets are being tested. A full understanding of chromatin structure and the mechanisms by which it is manipulated is therefore of vital importance.
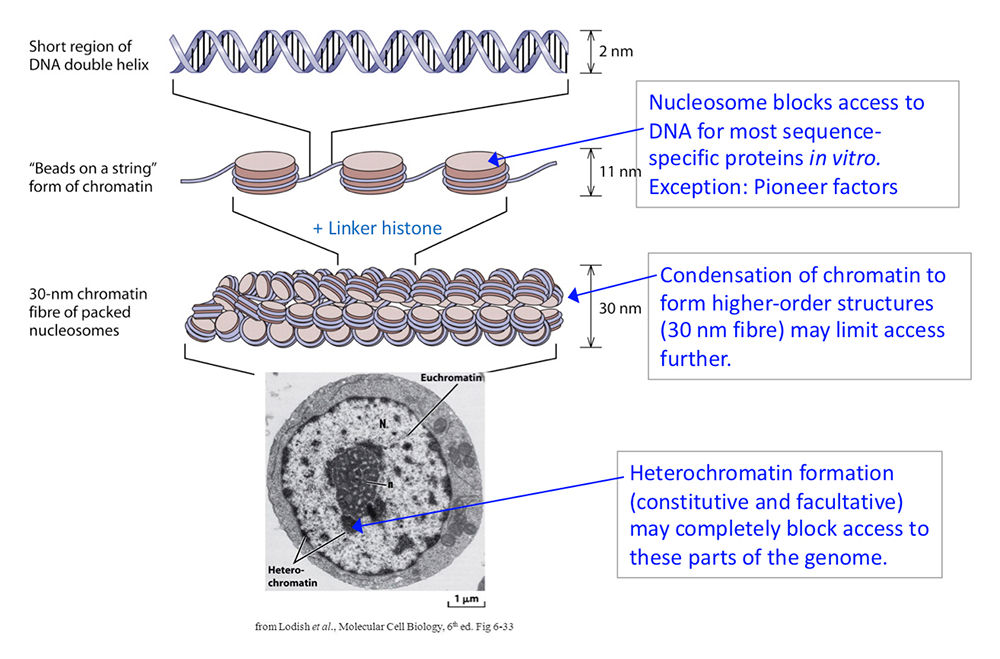
Figure 1. DNA packaging in the nucleus: to what extent does chromatin compaction limit access to the DNA?
DNA is packaged into the nucleus by histones. The basic structural subunit of chromatin is the nucleosome core, which contains about 147 bp of DNA wrapped nearly twice around a central octamer of core histones. Nucleosomes are regularly spaced along the DNA like beads on a string; the intervening DNA is called the linker DNA and is bound by linker histone (H1). The beads-on-a-string fiber spontaneously condenses into a heterogeneous fiber of about 30 nm width. Genomic regions rich in repetitive elements form constitutive heterochromatin in all cells, in which the chromatin fiber is even more condensed. Facultative heterochromatin is formed on genes that should be permanently silent in a specific differentiated cell type. Heterochromatin is densely packed and darkly staining in the electron micrograph shown here. Euchromatin is less condensed (light staining) and contains active genes. We are interested in determining to what extent chromatin limits DNA accessibility. Figure adapted from Chereji et al. Genome Res 2019;29:1985-1995.
Chromatin remodeling and DNA accessibility
Gene activation involves the recruitment of a set of factors to a promoter in response to appropriate signals, ultimately resulting in the formation of an initiation complex by RNA polymerase II and transcription, events that coincide with the removal of promoter nucleosomes to create a nucleosome-depleted region (NDR). This observation has led to the generally accepted model that promoter nucleosomes physically block transcript initiation, acting as repressors by preventing access to specific transcription factor binding sites. The nucleosome is a highly stable structure containing tightly wound DNA that is largely inaccessible to sequence-specific DNA–binding proteins. Activation occurs if sequence-specific ‘pioneer’ transcription factors are present (proteins that bind to nucleosomal sites with high affinity), and/or if ‘classical’ transcription factors, which are normally blocked by nucleosomes, recruit ATP–dependent chromatin remodelers to move or evict promoter nucleosomes, thus facilitating initiation complex formation.
The ATP–dependent chromatin remodelers variously move nucleosomes along DNA, remove the histones altogether, or form arrays of regularly spaced nucleosomes. Examples include the SWI/SNF and RSC complexes, which remodel nucleosomes on genes and at promoters, and the CHD and ISWI complexes, some of which are involved in determining nucleosome spacing. The INO80C complex is unusual because it has both properties. All these remodeling enzymes have human homologs.
Our activities this year were severely curtailed by the pandemic, but we were still able to complete two projects. In the first, we focused on understanding the roles of the two forms of the ISW1 complex in yeast [Reference 1]. Two different ATP–dependent nucleosome-spacing enzymes are required to establish regular arrays of phased nucleosomes near transcription start sites of yeast genes: Isw1 and Chd1. Cells lacking both Isw1 and Chd1 have extremely disrupted chromatin, such that the nucleosomes are poorly spaced and poorly positioned relative to the transcription start site. We had shown previously that such chromatin disorganization is at least partly attributable to a propensity in the double mutant to form close-packed dinucleosomes near the beginnings of genes (i.e., two nucleosomes jammed close together with no intervening linker DNA). Reaching a mechanistic understanding of the roles of the different remodelers is complicated by the fact that the Isw1 ATPase subunit occurs in two different remodeling complexes: ISW1a (composed of Isw1 and Ioc3) and ISW1b (composed of Isw1, Ioc2, and Ioc4). We constructed yeast strains with various combinations of deletions in the Ioc2/3/4 subunits and determined their chromatin organization using MNase-Seq, which provides a detailed nucleosome map for each strain. We discovered that ISW1b is primarily responsible for setting nucleosome spacing and resolving close-packed dinucleosomes, whereas ISW1a plays only a minor role. ISW1b and Chd1 make additive contributions to dinucleosome resolution, such that neither enzyme is capable of resolving all dinucleosomes on its own.
Given that the Ioc4 subunit of ISW1b binds preferentially to the histone H3-K36me3 mark, which is mediated by the Set2 H3-K36 methyltransferase and associated with active transcription, we tested whether loss of Set2 phenocopies loss of Ioc4. We found that dinucleosome levels are higher in cells lacking Set2, as is true for cells lacking Ioc4 (ISW1b), but, unlike ioc4-null cells, set2-null cells exhibit only a weak effect on nucleosome spacing. We propose that Set2–mediated H3-K36 trimethylation is important for ISW1b–mediated dinucleosome separation which, in turn, may be important for facilitating the passage of RNA polymerase II through the nucleosomes. We conclude that the nucleosome spacing and dinucleosome-resolving activities of ISW1b and Chd1 are critical for normal global chromatin organization, whereas ISW1a plays little or no role in chromatin organization at the global level. Thus, ISW1b is responsible for the effects of Isw1 on global chromatin structure, whereas the role of ISW1a is still unclear.
Global histone-surface accessibility in yeast indicates a uniformly loosely packed genome
Almost all methods designed to probe global chromatin structure measure DNA accessibility to nucleases (e.g. MNase-Seq and ATAC-Seq) or employ chemical crosslinking (e.g. ChIP-Seq and HiC). In a collaboration with Jeff Hayes, we developed a method to measure genome-wide accessibility of histone protein surfaces within nucleosomes by assessing reactivity of engineered cysteine residues with biotin-maleimide (BM), a thiol-specific reagent [Reference 2]. We determined the accessibility of three different histone residues by mutating them to cysteine in the appropriate yeast histone gene and then treating purified nuclei with BM. The chromatin is digested to mono-nucleosomes, streptavidin is used to purify modified nucleosomes, and the nucleosomal DNA is subjected to paired-end sequencing. Sequence data for modified nucleosomes are compared with data for input nucleosomes. In the first case, we measured the accessibility of the histone mutant H2B-S116C. The cysteine might be expected to be accessible, unless the chromatin is fully condensed, preventing BM modification. We found that it is generally accessible throughout the genome, suggesting that this histone surface is not usually obscured by tight nucleosome packing. In the second and third cases, we investigated the accessibility of cysteine residues buried inside the nucleosome, which might be exposed if the nucleosome is conformationally altered during transcription, as suggested by studies from other labs: H3-S102C, located at the H2A–H2B dimer/H3–H4 tetramer interface, and H3-A110C, located at the H3–H3 interface. However, we found that neither of these internal sites are significantly exposed. In conclusion, our data are consistent with a global, relatively decondensed chromatin structure, containing nucleosomes that do not generally undergo major conformational changes.
Role of the ARID1A subunit of the mouse SWI/SNF complex in gene regulation.
In a collaboration with Tom Owen-Hughes, we investigated the effects on gene regulation of depleting mouse cells of ARID1A, a critical subunit of the SWI/SNF ATP–dependent chromatin-remodeling complex. We found that loss of ARID1A results in a re-organization of chromatin structure around pluripotent transcription factor binding sites and a redistribution of the EP300 coactivator [Reference 3]. At enhancers, loss of EP300 correlates with reduced gene expression, whereas gain of EP300 correlates with increased gene expression. Surprisingly few genes are affected directly by loss of EP300, but cumulative indirect effects eventually phenocopy pre-malignancy.
Publications
- Eriksson PR, Clark DJ. The yeast ISW1b ATP-dependent chromatin remodeler is critical for nucleosome spacing and dinucleosome resolution. Sci Rep 2021;11:4195–4202.
- Marr LT, Ocampo J, Clark DJ, Hayes JJ. Global histone protein surface accessibility in yeast indicates a uniformly loosely packed genome with canonical nucleosomes. Epigenetics Chromatin 2021;14:5–15.
- Blümli S, Wiechens N, Wu M, Singh V, Gierlinski M, Schweikert G, Gilbert N, Naughton C, Sundaramoorthy R, Varghese J, Gourlay R, Soares R, Clark D, Owen-Hughes T. Acute depletion of the ARID1A subunit of SWI/SNF complexes reveals distinct pathways for activation and repression of transcription. Cell Reports 2021;37:109943.
Collaborators
- Harold Burgess, PhD, Section on Behavioral Neurogenetics, NICHD, Bethesda, MD
- Jeffrey J. Hayes, PhD, University of Rochester Medical Center, Rochester, NY
- Tom Owen-Hughes, PhD, University of Dundee, Dundee, United Kingdom
- Vasily M. Studitsky, PhD, Fox Chase Cancer Center, Philadelphia, PA
Contact
For more information, email clarkda@mail.nih.gov or visit http://clarklab.nichd.nih.gov.