Three-Dimensional Organization of the Genome as a Determinant of Cell-Fate Decisions

- Pedro Rocha, PhD, Head, Unit on Genome Structure and Regulation
- Nina Kopitchinski, Lab Manager
- Shreeta Chakraborty, PhD, Postdoctoral Fellow
- Joyce J. Thompson, PhD, Postdoctoral Fellow
- Zhenyu Zuo, PhD, Postdoctoral Fellow
- Dhanya Asokumar, BA, Postbaccalaureate Fellow
- Samuel Hernandez, BA, Postbaccalaureate Fellow
- Daniel Lee, BSc, Postbaccalaureate Fellow
Our lab seeks to understand cell-lineage differentiation, gene regulation, and how non-coding DNA elements and the 3D architecture of chromosomes contribute to such processes during development and disease. We are also interested in early mammalian development as a system in which to decipher how cells make lineage decisions and how gene-regulatory networks are established.
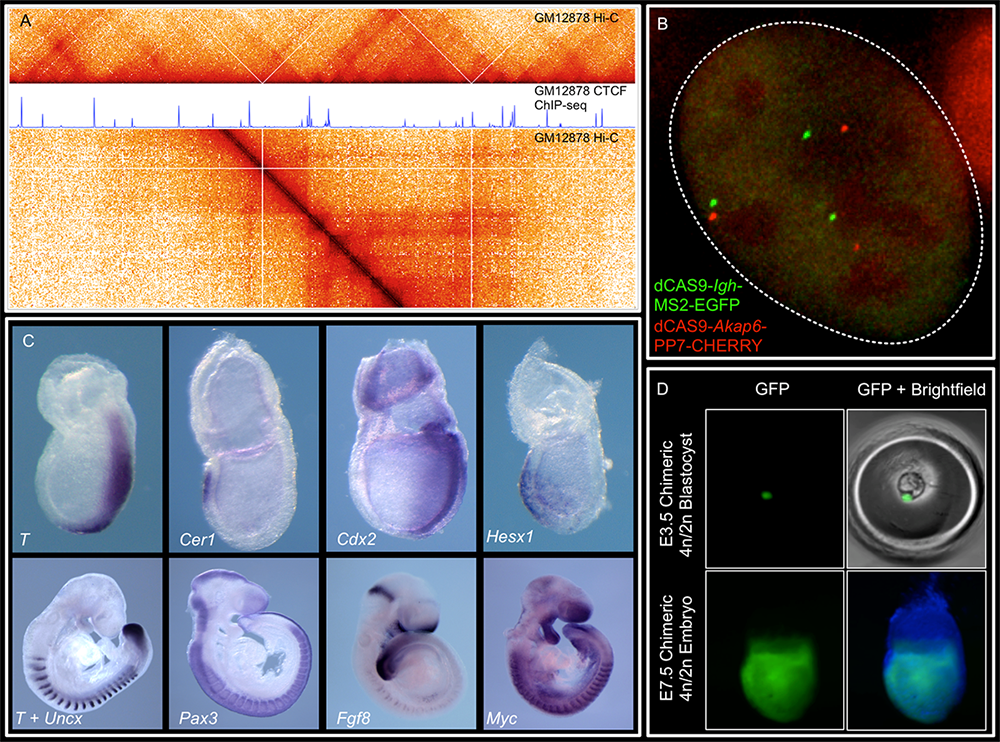
Figure 1. Representative image of the lab's research
We combine imaging techniques in both fixed and living cells with sequencing-based genomic techniques that assess DNA–DNA interactions.
A. Hi-C and CTCF ChIP-Seq of GM1278 cells, which allow characterization of chromatin structure and identification of binding sites of an important architectural protein.
B. dCAS9 MCP-EGFP and PCP-CHERRY live imaging of the Igh and Akap6 loci. The mouse embryo is an unparalleled system in mammalian biology for understanding how tissue-specific gene expression is achieved.
C. Whole-mount in situ hybridization for patterning markers in mid and late gastrulating embryos
D. Tetraploid aggregation with GFP (green fluorescent protein)–labeled ES (embryonic stem) cells allows generation of fully ES cell–derived embryos.
Eukaryotic cells need to deal with the biophysical constraints of packaging two meters of DNA inside a tiny nucleus (2–10 microns) and still retain the ability to access both its coding and non-coding elements to precisely orchestrate gene expression programs. Research over the past decade has begun to elucidate the mechanisms through which DNA condensation and organization in the nucleus are achieved. The results of such research suggest that the processes are tightly controlled and are themselves critical components of gene regulation. Our long-term goal is to understand how such processes occur in vivo and how their regulation dictates cell identity and cell-fate decisions in mammals.
To do so, we combine the robustness of mouse-genome editing and genetics with cutting-edge sequencing-based genomic techniques such as ATAC-Seq (assay for transposase-accessible chromatin using sequencing), ChIP-Seq (chromatin-immuno-precipitation DNA-sequencing), and Hi-C (high-throughput chromosome conformation capture technique), as well as live-imaging approaches. We believe that the early mouse embryo is an ideal model system in which to determine how nuclear architecture is regulated in the context of an organism and how that impacts cell behavior and identity.
Fertilization is the ultimate reprogramming experiment, where two highly differentiated cells (oocyte and sperm) fuse to form a zygote with totipotent potential. This involves a massive rearrangement of epigenetic modifications, both at the level of the DNA and of the histones, and the activity of many transcriptional regulators. Our studies aim to understand how 3D chromatin structures are established during this period and how they impact future developmental decisions.
Following fertilization and within a few cell divisions, the first cell lineages are established and different gene-expression programs are put into action. In mammals, the result is the formation of the blastocyst, a structure that contains three different cell types, each with a defined differentiation potential. The trophectoderm is responsible for forming the placenta, the primitive endoderm leads to the yolk sac, and the epiblast gives rise to all remaining embryonic tissues. We will build on decades of lineage-fate experiments and precisely characterized signaling pathways known to regulate early mouse development to understand the contribution of nuclear organization to gene regulation during these early cell fate decisions.
We are also interested in understanding not only how DNA organization impacts cell behavior, and ultimately animal development and health, but also the mechanisms through which DNA folding itself is established and regulated, and which proteins are involved in these processes. To broadly address such questions, we will employ several high-throughput technologies that we have established in the lab, in combination with genome-wide CRISPR screens. Ultimately, we will fully characterize in vivo candidates identified this way to stringently determine their impact on gene regulation during mammalian development.
Additional Funding
- DIR Scientific Director's Award
Publications
- Thompson JJ, Lee DJ, Mitra A, Frail S, Dale R, Rocha PP. Rapid redistribution and extensive binding of NANOG and GATA6 at shared regulatory elements underlie specification of divergent cell fates. bioRxiv 2021; https://doi.org/10.1101/2021.07.28.454132.
- Zuo Z, Rocha PP. Repetitive elements: different subtypes hint at distinct functions. Trends Genet 2021;36(6):385–387.
- Beck DB, Basar MA, Asmar AJ, Thompson JJ, Oda H, Uehara DT, Saida K, Pajusalu S, Talvik I, D'Souza P, Bodurtha J, Mu W, Baranano KW, Miyake N, Wang R, Kempers M, Tamada T, Nishimura Y, Okada S, Kosho T, Dale R, Mitra A, Macnamara E, Matsumoto N, Inazawa J, Walkiewicz M, Ounap K, Tifft CJ, Aksentijevich I, Kastner DL, Rocha PP, Werner A. Linkage-specific deubiquitylation by OTUD5 defines an embryonic pathway intolerant to genomic variation. Sci Adv 2021;7(4):eabe2116.
- Tottone L, Lancho O, Loh JW, Singh A, Kimura S, Roels J, Kuchmiy A, Strubbe S, Lawlor MA, da Silva-Diz V, Luo S, Gachet S, Garcia-Prieto CA, Hagelaar R, Esteller M, Meijerink JPP, Soulier J, Taghon T, Van Vlierberghe P, Mullighan CG, Khiabanian H, Rocha PP, Herranz D. A tumor suppressor enhancer of PTEN in T-cell development and leukemia. Blood Cancer Discov 2021;2(1):92–109.
- Rhodes C, Mitra A, Lee D, Lee D, Zhang Y, Thompson J, Rocha P, Dale R, Petros T. Single cell chromatin accessibility reveals regulatory elements and developmental trajectories in the embryonic forebrain. bioRxiv 2021; https://doi.org/10.1101/2021.03.21.436353.
- Payer LM, Steranka JP, Kryatova MS, Grillo G, Lupien M, Rocha PP, Burns KH. Alu insertion variants alter transcript levels. Genome Res 2021;31(12):2236–2248.
Collaborators
- Sevinc Ercan, PhD, New York University, New York, NY
- Stefan Feske, MD, New York University School of Medicine, New York, NY
- Daniel Herranz, PharmD, PhD, Cancer Institute of New Jersey, Rutgers University, New Brunswick, NJ
- Timothy Petros, PhD, Unit on Cellular and Molecular Neurodevelopment, NICHD
- Danny Reinberg, PhD, New York University School of Medicine, New York, NY
- Achim Werner, PhD, Stem Cell Biochemistry Unit, NIDCR, Bethesda, MD
Contact
For more information, email pedro.rocha@nih.gov or visit https://rochalab.nichd.nih.gov.