Regulatory Small RNAs and Small Proteins

- Gisela Storz, PhD, Head, Section on Environmental Gene Regulation
- Aixia Zhang, PhD, Staff Scientist
- Jordan J. Aoyama, MD, Postdoctoral Fellow
- Aisha Burton Okala, PhD, Postdoctoral Fellow
- Sahar Melamed, PhD, Postdoctoral Fellow
- Narumon Thongdee, PhD, Postdoctoral Fellow
- Lauren R. Walling, PhD, Postdoctoral Fellow
- Rilee D. Zeinert, PhD, Postdoctoral Fellow
- Aoshu Zhong, PhD, Postdoctoral Fellow
- Abigail M. Daniels, MS, Postbaccalaureate Fellow
- Katherine M. Klier, BS, Postbaccalaureate Fellow
- Kyle D. Rekedal, BS, Postbaccalaureate Fellow
- Tiara D. Tillis, BA, Postbaccalaureate Fellow
The group currently has two main interests: identification and characterization of small noncoding RNAs; and identification and characterization of small proteins of less than 50 amino acids. Both small RNAs and small proteins have been overlooked because they are not detected in biochemical assays, and the corresponding genes are missed by genome annotation and are poor targets for genetic approaches. However, both classes of small molecules are now being found to have important regulatory roles in organisms ranging from bacteria to humans.
Identification and characterization of small regulatory RNAs
During the past 20 years, we carried out several different systematic screens for small regulatory RNA genes in Escherichia coli, which showed that small RNAs are encoded by diverse loci, including sequences overlapping mRNAs [Reference 1]. The screens included computational searches for conservation of intergenic regions and direct detection after size selection or co-immunoprecipitation with the RNA–binding protein Hfq. Most recently, we have been using deep sequencing approaches to map the 5′ and 3′ ends of all transcripts to further extend our identification of small RNAs in a range of bacteria species [Reference 2]. The work uncovered several small RNAs encoded entirely within open reading frames (ORFs), including an RNA internal to an essential cell-division gene, which was found to have an independent function as a sponge that blocks the activity of another small RNA.
A major focus for the group was to elucidate the functions of the small RNAs that we and others had identified. Early on, we showed that the OxyS RNA, whose expression is induced in response to oxidative stress, acts to repress translation through limited base pairing with target mRNAs. We discovered that OxyS action is dependent on the Sm–like Hfq protein, which acts as a chaperone to facilitate OxyS RNA base pairing with its target mRNAs. We also explored the role of ProQ, a second RNA chaperone in E. coli and, by comparing the small RNA–mRNA interactomes by deep sequencing, we found that Hfq and ProQ have overlapping as well as competing roles in the cell [Reference 3]. In organisms that do not have Hfq or ProQ, other proteins such as KhpA and KhpB bind to small RNAs and may have similar chaperone roles [Reference 4].
It is clear that Hfq–binding small RNAs, which act through limited base pairing, are integral to many different stress responses in E. coli and other bacteria, as well as during the interaction between bacteria and bacteriophage [Reference 5]. For example, we showed that the Spot 42 RNA, whose levels are highest when glucose is present, plays a broad role in catabolite repression by directly repressing genes involved in central and secondary metabolism, redox balancing, and the consumption of diverse non-preferred carbon sources. Similarly, we discovered that a Sigma(E)–dependent small RNA, MicL, transcribed from a promoter located within the coding sequence of the cutC gene, represses synthesis of the lipoprotein Lpp, the most abundant protein in the cell, to oppose membrane stress. We found that the copper-sensitivity phenotype previously ascribed to inactivation of the cutC gene is in fact derived from the loss of MicL and elevated Lpp levels. As more and more small RNAs encoded by 5′ or 3′ UTRs or internal to coding sequences are being found, our observations regarding MicL raise the possibility that other phenotypes currently attributed to protein defects are attributable to deficiencies in unappreciated regulatory RNAs.
In addition to small RNAs that act via limited base pairing, we have been interested in regulatory RNAs that act by other mechanisms. For instance, early work showed that the 6S RNA binds to and modulates RNA polymerase by mimicking the structure of an open promoter. In another study, we discovered that the yybP-ykoY motif, a broadly conserved RNA structure motif found in the 5′ UTR of the mntP gene encoding a manganese exporter, directly binds to manganese, resulting in a conformation that liberates the ribosome-binding site.
Further studies to characterize other Hfq– and ProQ–binding RNAs and their physiological roles and evolution, as well as regulatory RNAs that act in ways other than base pairing, are ongoing.
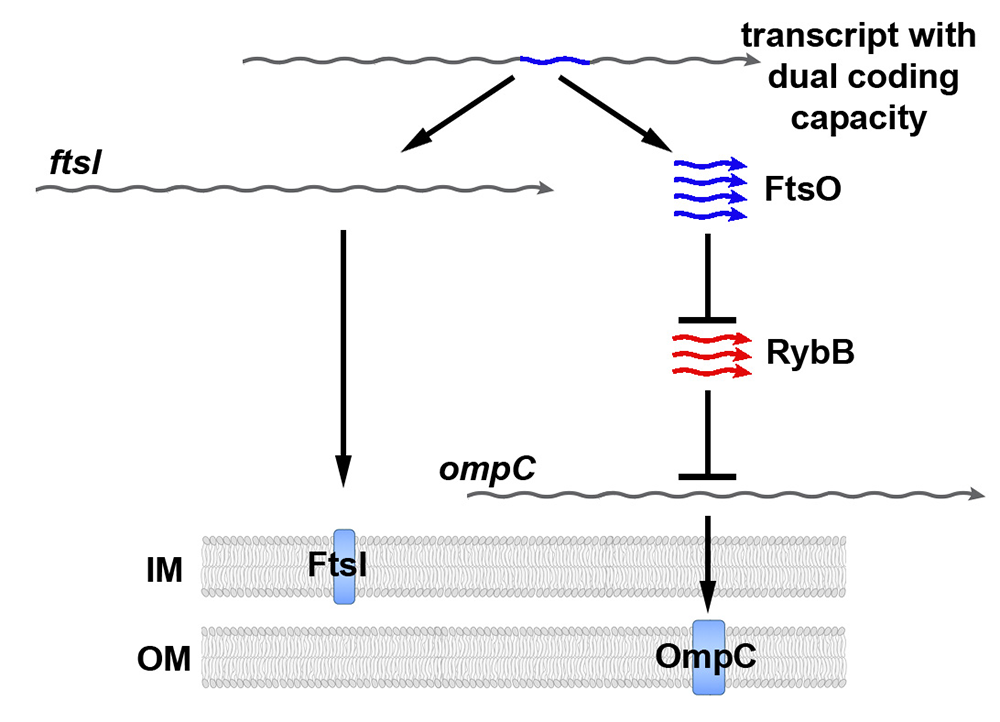
Figure 1. ORF–internal small RNA FtsO acts as a sponge of the RybB base-pairing small RNA.
Model showing how the same DNA sequence can encode two different gene products. The ftsI mRNA encodes the essential FtsI cell division protein found in the inner membrane (IM). The transcript also encodes the FtsO small RNA (blue), which blocks the activity of the RybB base-pairing small RNA (red). When not blocked by FtsO, RybB down-regulates the synthesis of outer membrane (OM) porins such as OmpC.
Identification and characterization of small proteins
In our genome-wide screens for small RNAs, we found that a number of short RNAs actually encode small proteins. The correct annotation of the smallest proteins is one of the biggest challenges of genome annotation, and there is limited evidence that proteins are synthesized from annotated and predicted short ORFs. Although these proteins have largely been missed, the few small proteins that have been studied in detail in bacterial and mammalian cells have been shown to have important functions in regulation, signaling, and cellular defenses [Reference 6]. We thus established a project to identify and characterize proteins of less than 50 amino acids.
We first used sequence conservation and ribosome binding-site models to predict genes encoding small proteins of 16–50 amino acids in the intergenic regions of the model E. coli genome. We tested expression of these predicted as well as previously annotated small proteins by integrating the sequential peptide affinity tag directly upstream of the stop codon on the chromosome and assaying for synthesis using immunoblot assays. This approach confirmed that 20 previously annotated and 18 newly discovered proteins of 16–50 amino acids are synthesized. We also carried out a complementary approach based on genome-wide ribosome profiling of ribosomes arrested in start codons to identify many additional candidates; the synthesis of 38 of these small proteins was confirmed by chromosomal tagging. These studies, together with the work of others, documented that E. coli synthesize over 150 small proteins [Hemm MR, Weaver J, Storz G. EcoSal Plus 2020; 9:doi:10.1128/ecosalplus.ESP-0031-2019].
Many of the initially discovered proteins were predicted to consist of a single transmembrane alpha-helix and, by biochemical fractionation, were found to be in the inner membrane. Interestingly, assays of topology-reporter fusions and strains with defects in membrane-insertion proteins revealed that, despite their diminutive size, small membrane proteins display considerable diversity in topology and insertion pathways. Additionally, systematic assays of the levels of tagged derivatives showed that many small proteins accumulate under specific growth conditions or after exposure to stress. We also generated and screened bar-coded null mutants and identified small proteins required for resistance to cell-envelope stress and acid shock.
We now are using the tagged derivatives and information about synthesis and subcellular localization, and are employing many of the approaches the group has used to characterize the functions of small regulatory RNAs to elucidate the functions of the small proteins. The combined approaches are beginning to yield insights into how the small proteins are acting in E. coli. For example, we discovered the 49–amino acid inner membrane protein AcrZ (formerly named YbhT), whose synthesis is increased in response to noxious compounds such as antibiotics and oxidizing agents, associates with the AcrAB-TolC multidrug efflux pump, which confers resistance to a wide variety of antibiotics and other compounds. Co-purification experiments, two-hybrid assays, and suppressor mutations showed that AcrZ interacts directly with the inner-membrane protein AcrB. Mutants lacking AcrZ are sensitive to many, but not all, the antibiotics transported by AcrAB-TolC. The differential antibiotic sensitivity suggests that AcrZ enhances the ability of the AcrAB-TolC pump to export certain classes of substrates. Detailed cryo-EM and mutational studies recently showed that AcrZ and cardiolipin cooperate to allosterically modulate AcrB activity [Du D et al., Structure 2020; 28:625-634].
Two small proteins act to increase the intracellular levels of manganese and magnesium. We found that synthesis of a 42–amino acid protein, now denoted MntS, is increased under low magnesium conditions, which eliminates repression by MntR. The lack of MntS leads to decreased activities of manganese-dependent enzymes under manganese-poor conditions, while overproduction of MntS leads to very high intracellular manganese and bacteriostasis under manganese-rich conditions. These and other phenotypes led us to propose that MntS modulates intracellular manganese levels, possibly by inhibiting the manganese exporter MntP. We also showed that the 31–amino acid inner membrane protein MgtS (formerly denoted YneM), whose synthesis is induced by very low magnesium in a PhoPQ–dependent manner, acts to increase intracellular magnesium and maintain cell integrity upon magnesium depletion. Upon development of a functional tagged derivative of MgtS, we found that MgtS interacts with MgtA to increase the levels of this P-type ATPase magnesium transporter under magnesium-limiting conditions. Correspondingly, the effects of MgtS upon magnesium limitation are lost in an mgtA mutant, and MgtA overexpression can suppress the mgtS phenotype. MgtS stabilization of MgtA provides an additional layer of regulation of this tightly controlled magnesium transporter. Most recently we found that MgtS also interacts with and modulates the activity of a second protein, the PitA cation-phosphate symporter, to further increase intracellular magnesium levels.
A limited number of transcripts encoding both a small protein and possessing base-pairing activity have been identified and denoted dual-function RNAs. However, given that few have been characterized, little is known about the interplay between the two regulatory functions. Interestingly, MgtS is encoded differently from the MgrR small regulatory RNA, which is also important for bacterial adaptation to low magnesium. To investigate the competition between protein-coding and base-pairing activities, we constructed synthetic dual-function RNAs comprised MgrR and MgtS [Aoyama JJ, Raina M, Storz G. J Bacteriol 2021; 204(1):JB0034521]. These constructs allowed us to probe how the organization of components and the distance between the coding and base-pairing sequences contribute to the proper function of both activities of a dual-function RNA. By understanding the features of natural and synthetic dual-function RNAs, future synthetic molecules can be designed to maximize their regulatory impact.
This work, together with our ongoing studies of other small proteins and related findings by others in eukaryotic cells, supports our hypothesis that small proteins are an overlooked class of regulators.
Additonal Funding
- NICHD Early Career Award
- NIGMS Postdoctoral Research Associate (PRAT) Program
Publications
- Adams PP, Storz G. Prevalence of small base-pairing RNAs derived from diverse genomic loci. Biochim Biophys Acta Gene Regul Mech 2020;1863:194524.
- Adams PP, Baniulyte G, Esnault C, Chegireddy K, Singh N, Monge M, Dale RK, Storz G, Wade JT. Regulatory roles of Escherichia coli 5´ UTR and ORF-internal RNAs detected by 3´ end mapping. eLife 2020;10:e62438.
- Melamed S, Adams PP, Zhang A, Zhang H, Storz G. RNA-RNA interactomes of ProQ and Hfq reveal overlapping and competing roles. Mol Cell 2020;77:411–425.e7.
- Olejniczak M, Jiang X, Basczok MM, Storz G. KH-domain proteins: another family of bacterial RNA matchmakers? Mol Microbiol 2022;117(1):10–19.
- Hör J, Matera G, Vogel J, Gottesman S, Storz G. Trans-acting small RNAs and their effects on gene expression in Escherichia coli and Salmonella enterica. EcoSal Plus 2020;ESP-0030-2019.
- Wu Orr M, Mao Y, Storz G, Qian SB. Alternative ORFs and small ORFs: shedding light on the dark proteome. Nucleic Acids Res 2020;48(3):1029–1042.
Collaborators
- Ryan K. Dale, PhD, Bioinformatics and Scientific Programming Core, NICHD, Bethesda, MD
- Susan Gottesman, PhD, Laboratory of Molecular Biology, Center for Cancer Research, NCI, Bethesda, MD
- Xiaofang Jiang, PhD, National Library of Medicine, NIH, Bethesda, MD
- Dijun Du, PhD, School of Life Science and Technology, ShanghaiTech University, Pudong, China
- Todd Gray, PhD, Wadsworth Center, New York State Department of Health, Albany, NY
- Matthew R. Hemm, PhD, Department of Biological Sciences, Towson University, Towson, MD
- Syma Khalid, PhD, University of Southampton, Southampton, United Kingdom
- Ben F. Luisi, PhD, University of Cambridge, Cambridge, United Kingdom
- Mikolaj Olejniczak, PhD, Institute of Molecular Biology and Biotechnology, Adam Mickiewicz University, Poznan, Poland
- Kai Papenfort, PhD, Institute of Microbiology, Friedrich-Schiller-Universität, Jena, Germany
- Shu-Bing Qian, PhD, Cornell University, Ithaca, NY
- Jörg Vogel, Dr rer nat, Institute of Molecular Infection Biology, Universität Würzburg, Würzburg, Germany
- Joseph T. Wade, PhD, Wadsworth Center, New York State Department of Health, Albany, NY
- Henry Zhang, PhD, Bioinformatics and Scientific Programming Core, NICHD, Bethesda, MD
Contact
For more information, email storz@helix.nih.gov or visit https://storz.nichd.nih.gov.