The Biological Impact of Transposable Elements

- Henry L. Levin, PhD, Head, Section on Eukaryotic Transposable Elements
- Angela Atwood-Moore, BA, Senior Research Assistant
- Abhishek Anand, PhD, Postdoctoral Fellow
- Paul Atkins, PhD, Postdoctoral Fellow
- Hyo Won Ahn, PhD, Visiting Fellow
- Feng Li, PhD, Visiting Fellow
- Rakesh Pathak, PhD, Visiting Fellow
- Kyla Roland, BA, Postbaccalaureate Fellow
Long terminal repeat (LTR)–retrotransposons constitute a significant fraction of eukaryotic genomes and have produced a large share of their genetic diversity. Their infectious counterparts, the retroviruses, are wide-spread pathogens of vertebrates, which evolved over 450 million years ago. LTR–retrotransposons and retroviruses propagate through a unique cycle in which an RNA intermediate is reverse-transcribed into cDNA copies that are inserted into host chromosomes by integrases (IN). The integration of cDNA results in dysregulation and outbreaks of neoplastic disease in a wide range of species, including salmon and koala bear, and in the case of the worldwide pandemic of AIDS, integration of HIV-1 results in acute loss of immune cells and tragic rates of mortality. Potent inhibitors of IN–strand transfer activity are a frontline component of antiretroviral treatment of AIDS patients, confirming that integration is a central feature of HIV-1 replication.
Despite the central role of integration, important questions remain about residual infection that occurs in the absence of IN activity. Mutations in HIV-1 IN of the catalytic residues produce residual infectious titer, typically with a 3 to 4-log decrease. However, in continuous cultures of HIV-1 lacking IN activity, insertion efficiency can be as high as 0.2–0.8% of the wild-type virus. Consistent with the studies of HIV-1, a distantly related retrovirus, murine leukemia virus (MLV), also possesses residual insertion activity when IN is inactivated. Such results indicate that retroviruses possess a secondary, IN–independent pathway, which incorporates viral DNA into the host genome. Given that IN–independent infections may compromise the treatment of HIV-1 patients with IN inhibitors, it is critical to identify the nature of this pathway.
Identification of an integrase-independent pathway of retrotransposition
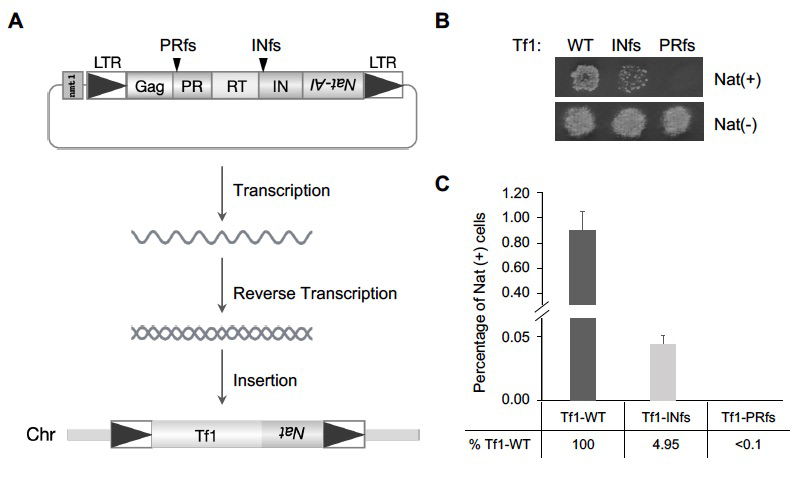
As a result of their structural similarities, LTR retrotransposons have been widely used as important models for studying the pathogeny of retroviruses. Tf1 and Tf2 are extensively characterized LTR retrotransposons with high integration activity in Schizosaccharomyces pombe [Reference 1]. We found that Tf1 retains 5% insertion activity in the absence of IN, which allowed us to study the mechanistic underpinnings of the process (Figure 1). With high-throughput sequencing, we found that insertions of Tf1 lacking IN (Tf1-INfs) occurred at sites that possessed homology to the primer-binding site (PBS) and poly purine tract (PPT), whose sequences serve as RNA primers of reverse transcription and that are copied into single stranded DNAs at the 3′ ends of the cDNA. Notably, we found that in previously published data of HIV-1 lacking IN activity insertions sites can have strong similarity to the PBS, indicating that this process maybe widespread among retroviruses.
Additional analysis revealed that a substantial fraction of Tf1–INfs insertions occurred adjacent to pre-existing retrotransposons, resulting in tandem structures that expand to serve as reservoirs of active elements. A genetic screen revealed that IN–independent insertions were mediated by Rad52 (a member of the homologous recombination pathway, important for maintenance of genome integrity). Mutations in rad52 showed that the Tf1 insertions result from single-strand annealing (SSA), a non-canonical form of homologous recombination mediated by Rad52 that is independent of Rad51. Surprisingly, we discovered that wild-type Tf1 can switch from IN–dependent to this IN–independent pathway of insertion depending on culture conditions. Taken together, we demonstrated that there are two efficient insertion pathways of cDNA, one relying on IN while the other is IN–independent but requires Rad52–mediated SSA.
Figure 1. Tf1 insertion takes place in the absence of integrase.
A. The diagram shows the strategy of monitoring Tf1 retrotransposition. A drug-resistant gene, nat, with artificial intron (nat-AI) is introduced into Tf1, and the integration of Tf1 into host chromosomes allows cells to grow on plates containing Nat. The black arrows indicate the frame shift (fs) sites of PR and IN respectively. LTR: long terminal repeat; PR: protease; RT: reverse transcriptase; IN: integrase; WT: wild-type.
B. Growth phenotypes of Tf1-WT, Tf1-INfs, and Tf1-PRfs on medium containing Nat after inducing Tf1 expression.
C. Quantitative transposition analysis Tf1-WT, Tf1-INfs, and Tf1-PRfs.
Dense transposon integration reveals that essential cleavage and polyadenylation factors promote heterochromatin formation.
In eukaryotes, the assembly of DNA into highly condensed heterochromatin is critical for a broad range of functions related to genome integrity. The methylation of histone H3 on lysine 9 (H3K9me) is central to the formation of heterochromatin by creating binding sites for a range of chromatin proteins important for silencing transposable elements, chromosome segregation, and epigenetic inheritance. Used extensively for this purpose, S. pombe is an excellent model in which to study the molecular mechanisms that generate and regulate heterochromatin. Centromeres, subtelomeres, and the mating-type region are packaged into constitutive heterochromatin, while meiosis genes are silenced by facultative heterochromatin until cells are starved of nitrogen. Importantly, Clr4, the H3K9–specific histone methyltransferase, is recruited to heterochromatin regions by several mechanisms. Constitutive heterochromatin results from RNAi factors that include the Ago1–containing, RNA–induced transcriptional silencing complex (RITS). Facultative heterochromatin at meiosis genes is independent of RNAi and relies on the RNA elimination (i.e., degradation) factors Red1 and Mmi1 and on the nuclear exosome. However, gaps exist in our understanding of how RNA elimination generates heterochromatin. A new approach to identifying gene function is the high-throughput sequencing of integration profiles, also known as Tn-Seq, which identifies genes important for growth under selective conditions. Genes necessary to sustain growth under a specific condition do not tolerate insertions in that condition. Tn-Seq has been applied to identify pathogenic genes in bacteria. However, we were the first to develop the method for a eukaryote; we developed a method for identifying essential genes in yeast, and others have subsequently applied the strategy to single-cell eukaryotes [References 2–5].
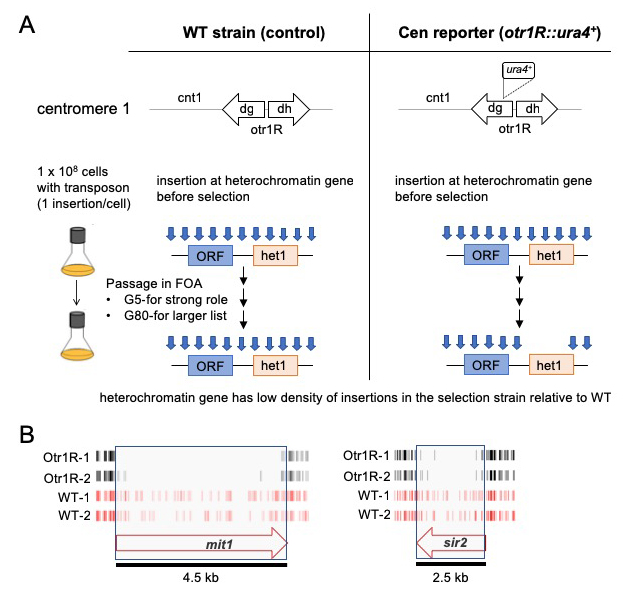
With the goal of identifying novel factors important for heterochromatin, we produced dense profiles of integrations using the Hermes transposable element and a silencing reporter (ura4) positioned in the outer repeats of centromere 1 [Reference 2]. Inserts that disrupted genes important for heterochromatin activated ura4, and thus the cells were unable to grow when passaged in 5-fluoroorotic acid (FOA) (Figure 2A). Genes with established roles in heterochromatin assembly had significantly fewer insertions in cells with the centromere reporter otr1R::ura4 than in cells lacking the reporter (Figure 2B). The list of candidates consisted of a total of 199 genes and, importantly, 65 are known to be essential for viability. These essential genes were candidates because they tolerated many insertions in their 3′ sequences that reduced heterochromatin but not viability. The high number of essential genes is significant in that most proteins found to be important for heterochromatin are identified in screens of deletion strains that cannot include essential genes. The 199 candidates showed highly significant enrichments for functions in silencing at centromere outer repeats and included all four factors that produce siRNA.
Figure 2. Dense maps of transposable element integration identify genes important for heterochromatin at centromere repeats.
A. Single insertions of the transposable element Hermes were generated in cells with WT cen1 and cen1 otr1R::ura4. Cultures were passaged in 5-fluoroorotic acid (FOA) for 5 or 80 generations. Cells with insertions in heterochromatin genes (het1) express ura4 and cannot grow in FOA. After growth on FOA fewer insertions were detected in het genes in cells with cen1 otr1R::ura4.
B. Genes involved in forming centromere heterochromatin such as mit1 and sir2 had fewer inserts in cells with the cen1 otr1R::ura4 (black, dupl. libraries) than cells with WT cen1 (red, dupl. libraries).
We identified other RNA–processing factors that were not previously linked to heterochromatin structure. Strikingly, four of the RNA–processing candidates form an interaction module of the canonical mRNA polyadenylation factor and the cleavage factor CPF, as predicted from highly homologous proteins in S. cerevisiae. To determine whether polyadenylation and cleavage contribute to heterochromatin structure at the centromere repeats, we focused on the function of Iss1, a subunit of CPF. We generated a C-terminal truncation of Iss1 (Iss1-deltaC) by removing 38 amino acids that, based on the Hermes insertions, were not important for viability. Iss1-deltaC showed no growth restriction on nonselective medium but exhibited a heterochromatin defect, as demonstrated by growth in the absence of uracil and reduced levels of H3K9 dimethylation (H3K9me2) at otr1R::ura4. The results demonstrated that the Hermes screen correctly identified Iss1 as important for heterochromatin structure at the otr1R::ura4 reporter. Interestingly, we found that Iss1 contributes to the heterochromatin of centromere repeats in cells that lack the otr1R::ura4 reporter but, in this case, the contribution to H3K9me2 was only observed when the RNAi pathway was disabled by deletion of ago1. This role at the outer centromere repeats is therefore independent or redundant with RNAi.
We expanded our study of the Iss1-deltaC mutation to evaluate changes in expression and transcription termination genome-wide. RNA-Seq data revealed that Iss1-deltaC did not significantly impact canonical transcription termination, but 73 genes were found to have higher expression. Importantly, the genes overlapped significantly with genes upregulated in cells lacking Rrp6, the 3′-5′ exonuclease subunit of the nuclear exosome. As a key subunit of the nuclear exosome, Rrp6 plays an important role in RNA surveillance in the degradation of meiotic transcripts expressed during vegetative growth and the resulting formation of heterochromatin at these genes. The elimination of meiotic mRNAs depends on the RNA–binding protein Mmi1 to bind to the determinant of selective removal (DSR) sequence in order to recruit the exosome. Our co-immunoprecipitation experiments revealed that Iss1 interacted with Rrp6, Mmi1, and the polyA polymerase Pla1, indicating that Iss1 is associated with this network of elimination factors. Significantly, the interaction with Mmi1 was disrupted by the Iss1-deltaC mutation, a mutation that greatly reduced H3K9me2 at meiotic genes. We tested whether Iss1 plays a direct role in the heterochromatin of meiotic genes by performing ChIP-Seq of Iss1-FLAG. While a subset of Iss1–bound genes was highly expressed and associated with the canonical function of Iss1 in mRNA termination, most Iss1–bound peaks showed a strong correlation with genes regulated by RNA elimination and heterochromatin. Importantly, the iss1-deltaC mutation caused significant increases in the RNA levels of these genes. Taken together, our studies of RNA levels, Iss1 association with chromatin, and H3K9me2 indicate that Iss1 plays a direct role in the formation of heterochromatin at meiotic genes. Our application of Hermes profiles to identify genes important for heterochromatin formation demonstrates the significance of the approach, especially given that we were able to identify large numbers of essential genes, a result not obtainable with other screens.
Retrotransposon insertions associated with risk of neurologic and psychiatric diseases
Neurologic and psychiatric disorders affect 25% of the world population. Given the complexity of the mammalian nervous system, the genetic and cellular etiology of such diseases remains largely unclear. Progress in genetic methodology has provided the potential to identify mechanisms that underlie the diseases. One approach that has successfully identified important disease loci is genome-wide association studies (GWAS). However, in the cases of neurologic and major psychiatric disorders, GWAS have identified large numbers of loci, each associated with small increases in risk. Importantly, there is extensive overlap of the loci that contribute to major psychiatric disorders, indicating that related molecular mechanisms may underlie distinct clinical phenotypes.
Single-nucleotide polymorphisms (SNPs) identified by GWAS with the highest disease association
Trait associated SNPs (TASs) are genetic tags identifying a genomic region that contains the causal mutation(s) leading to the disease risk. Limits on the design of GWAS typically prevent such studies from identifying causal gene alleles. Determining causal variants remains the most challenging and rate-limiting, but also the most important step in defining the genetic architecture of diseases. The vast majority of GWAS TASs lie in intergenic or intronic regions and therefore do not alter coding sequence. For such SNPs to be causal they would likely have regulatory effects on transcription. Structural variants such as rearrangements, copy number variants, and transposable element (TE) insertions constitute a substantial and disproportionately large fraction of the genetic variants found to alter gene expression.
In humans, the dominant families of TEs are long interspersed element-1 (LINE-1 or L1) and Alu elements, which are short interspersed elements (SINEs) and are mobilized by L1. TEs alter gene expression particularly easily because they have evolved various sequences that act on enhancers. Given that TEs make up approximately 45% of the human genome, it is not surprising that their regulatory features are abundant sources of tissue-specific promoter activity.
Relatively recent TE insertions can proliferate in the population and become common alleles. The 1000 Genomes Project described genetic variation of diverse human populations by sequencing whole genomes of 2,504 individuals. The extensive survey of genetic variation detected 17,000 polymorphic insertions of TEs, which have the potential to alter gene expression and affect common-disease risk. Some TEs have been implicated at disease loci detected by GWAS.
Given the difficulty in identifying genetic variants responsible for neurologic and psychiatric disorders and the regulatory capacity of TEs, we tested whether polymorphic TEs are potential causative variants of such diseases. We analyzed 593 GWAS of neurologic and psychiatric diseases, which in total reported 753 TASs. From the 17,000 polymorphic TEs, we found that 76 were in linkage disequilibrium (LD) with TASs, indicating that the TEs were among the variants with the potential to be causative. We extended our analysis by evaluating each candidate TE for a role in altering expression of proximal genes. In one approach we determined whether polymorphic TEs could disrupt regulatory sequences, as annotated with the epigenomic data of the NIH Roadmap Epigenomics Consortium. Ten of the TE candidates were located in regions of chromatin with active regulatory function in neurologic tissues. We also tested whether the polymorphic TEs were significantly associated with altered expression of proximal genes. By analyzing multi-tissue expression data from GTEx (Genotype-Tissue Expression project), we found that 31 of the TASs linked to TEs were expression-quantitative trait loci (eQTLs, loci that seek to identify genetic variants that affect the expression of one or more genes) for adjacent genes, showing correlation with altered expression within regions of the brain. These expression data, together with epigenetic and eQTL analyses, indicate that polymorphic TE insertions are important candidates for causing disease risk for Parkinson's disease, schizophrenia, and amyotrophic lateral sclerosis, on par with other variants at these loci.
Additional Funding
- FY2021 Office of AIDS Research Innovative Funds Program
- NICHD Distinguished Scholars Program
Publications
- Esnault C, Lee M, Ham C, Levin HL. Transposable element insertion in fission yeast drives adaptation to environmental stress. Genome Res 2019;29:85–95.
- Lee SY, Hung S, Esnault C, Pathak R, Johnson K, Bankole O, Yamashita A, Zhang H, Levin HL. Dense transposon integration reveals essential cleavage and polyadenylation factors promote heterochromatin formation. Cell Rep 2020;30:2686–2698.
- Grech L, Jeffares DC, Sadée CY, Rodríguez-López M, Bitton DA, Hoti M, Biagosch C, Aravani D, Speekenbrink M, Illingworth CJR, Schiffer PH, Pidoux AL, Tong P, Tallada VA, Allshire R, Levin HL, Bähler J. Fitness landscape of the fission yeast genome. Mol Biol Evol 2019;36:1612–1623.
- van Opijnen T, Levin HL. Transposon insertion sequencing, a global measure of gene function. Annu Rev Genet 2020;54:337–365.
- Li F, Hung S, Esnault C, Levin HL. A protocol for transposon insertion sequencing in Schizosaccharomyces pombe to identify factors that maintain heterochromatin. STAR Protoc 2021;2:100392.
Collaborators
- Jürg Bähler, PhD, University College London, London, United Kingdom
- Shiv Grewel, PhD, Laboratory of Biochemistry and Molecular Biology, NCI, Bethesda, MD
- Stephen Hughes, PhD, Retroviral Replication Laboratory, HIV Drug Resistance Program, NCI, Frederick, MD
- Mamuka Kvaratskhelia, PhD, Ohio State University, Columbus, OH
- Matthew Plumb, BS, Ohio State University, Columbus, OH
- Akira Yamashita, PhD, National Institute for Basic Biology, Okazaki, Japan
Contact
For more information, email henry_levin@nih.gov or visit https://sete.nichd.nih.gov.