Receptors and Actions of Peptide Hormones and Regulatory Proteins in Endocrine Mechanisms

- Maria L. Dufau, MD, PhD, Head, Section on Molecular Endocrinology
- Raghuveer Kavarthapu, PhD, Staff Fellow
- Muruganath Kumar Raju, PhD, Postdoctoral Fellow
- Rajakumar Anbazhagan, PhD, Visiting Fellow
The Section investigates the regulatory mechanism(s) involved in the progression of spermatogenesis and the control of Leydig cell function. We investigate novel gonadotropin-regulated genes relevant to the progression of testicular gametogenesis, Leydig cell function, and other endocrine processes. Our studies concentrate on the function and regulation of the gonadotropin-regulated testicular RNA helicase (GRTH/DDX25), a member of the DEAD–box family of RNA helicases discovered, cloned, and characterized in our laboratory, which is essential for the progress of spermatogenesis [Reference 1]. The various functions of GRTH/DDX25 provide fertile ground for the development of a non-hormonal male contraceptive.
Gonadotropin-regulated testicular RNA helicase (GRTH)
GRTH is present in Leydig cells (LCs) and meiotic (pachytene spermatocytes) and haploid germ cells (round and elongated spermatids). Male null mice lacking GRTH are sterile owing to azoospermia resulting from the failure of round spermatids to elongate. We demonstrated GRTH's participation in the nuclear export/transport of specific mRNAs, the structural integrity of the chromatoid body (CB, an organelle prevalent in round spermatids [RS] and found to be key for the progress of spermatogenesis), storage/processing of relevant mRNAs, and their transit/association to the actively translating polyribosomes, where it may regulate translational initiation of genes. GRTH is the only DEAD-box family member regulated by hormones. GRTH transcription is stimulated in LCs by LH (luteinizing hormone)/cAMP through direct actions of the androgen (A)/A receptor (AR) (autocrine), and in germ cells in paracrine fashion through the AR in Sertoli cells. The upstream region of the GRTH gene directs its expression in germ cells and downstream in LCs. Through these regions, A/AR exerts its direct (endogenous) regulation of the GRTH gene in the LC, and indirectly in germ cells.
We identified functional binding sites for the germ-cell nuclear factor (GCNF), which are present in RS and spermatocytes (SP), and its regulation by A/AR in the distal region of the GRTH gene, operative selectively in RS. Current knowledge indicates actions of A on GCNF cell–specific regulation of GRTH expression in germ cells (RS). Also, GRTH exerts negative autocrine regulation of GCNF, linking A actions to germ cells through GCNF as an A–regulated trans-factor that controls transcription/expression of GRTH (Kavarthapu & Dufau, Mol Endocrinol 2015;29:1792). These findings provide a connection between androgen action and two relevant germ-cell genes (GRTH and GCNF), which are essential for the progress of spermatogenesis, and we established their regulatory interrelationship.
Our early studies revealed that when the missense mutation of R to H at aa 242 of GRTH, found in 5.8% of 100 azoospermic patients, was transfected in COS1 cells, it caused loss of the 61 kDa phospho-species (pGRTH) with preservation of the 52 kDa non-phospho form (Tsai-Morris et al., Mol Human Reprod 2007;13:887). The finding provided an avenue to elucidate the function of pGRTH in spermatogenesis. We generated humanized mutant GRTH knock-in (KI) mice. The mutant mice are sterile, with reduction on testicular size, they lack sperm with arrest at step 8 of round spermatids (RS), and exhibit complete loss of the pGRTH species with preservation of the nuclear 52 kDa form [Reference 2]. The mouse model allows us to study the biological/biochemical functions of the cytoplasmic pGRTH. In KI mice, the nuclear export/transport and functions of GRTH are preserved (i.e., mRNA export), while the cytoplasmic functions, including shuttling of messages, storage in the CB, and translational events all requiring pGRTH, are absent. We observed a marked reduction in the size of the CB in RS and lack of pGRTH in the CBs. Germ-cell apoptosis was present in pachytene spermatocytes (PS) and in RS. In contrast to KO, KI showed no changes in miRNA biosynthesis, excluding participation of pGRTH as a transcriptional regulator of the microprocessor complex (which consists of Drosha, a miRNA–processing enzyme and its cofactor the RNA–binding protein DCGR), affecting pri-miRNA (primary microRNA) formation and indicative of the participation of non-phospho GRTH in such processes. In KI mice there is loss of chromatin-remodeling and related proteins, including TP2 (transition protein 2, a testis-specific DNA–condensing protein), PRM2 (a protamine that facilitates DNA condensation), and TSSK6 (a testis-specific serine/threonone-protein kinase, which is involved in the compaction of DNA during spermatogenesis). Significant reductions in their mRNA and half-lives indicate that their association with pGRTH in the cytoplasm protects these mRNAs from degradation. Also, our work showed that pGRTH stimulates TP2 translation in a 3′UTR–dependent manner.
In recent studies, we elucidated the GRTH phospho-site at a threonine (T239), which is structurally adjacent to the mutant site found in patients (R242H). Molecular modelling of the phospho-site based on the RecA domain 1 of the DDX9 crystal structure pointed to the amino acids that formed the GRTH/protein kinase alpha (PKA) interface, solvent accessibility, and H-bonding. These include, in addition to the core residues T239 and R242, amino acids E165, K240, and D237 [Reference 3]. We demonstrated the relevance of these residues by disrupting amino acids using site-directed mutagenesis (single or double mutations), which caused reduction or abolition of the pGRTH at T239. The pGRTH form is the cytoplasmic species that was demonstrated as essential for the progress of spermatogenesis beyond step 8 of RS and for viable sperm formation. It is important to note that the deleterious effects on GRTH phosphorylation caused by the mutations did not result from changes in PKA–catalytic binding affinity but rather in consequential structural changes that can affect PKA catalytic efficiency. Studies based on the abolition of the phospho-form provide the basis for drug design, such as for discovery of a reversible chemical inhibitor for use as male contraceptive.
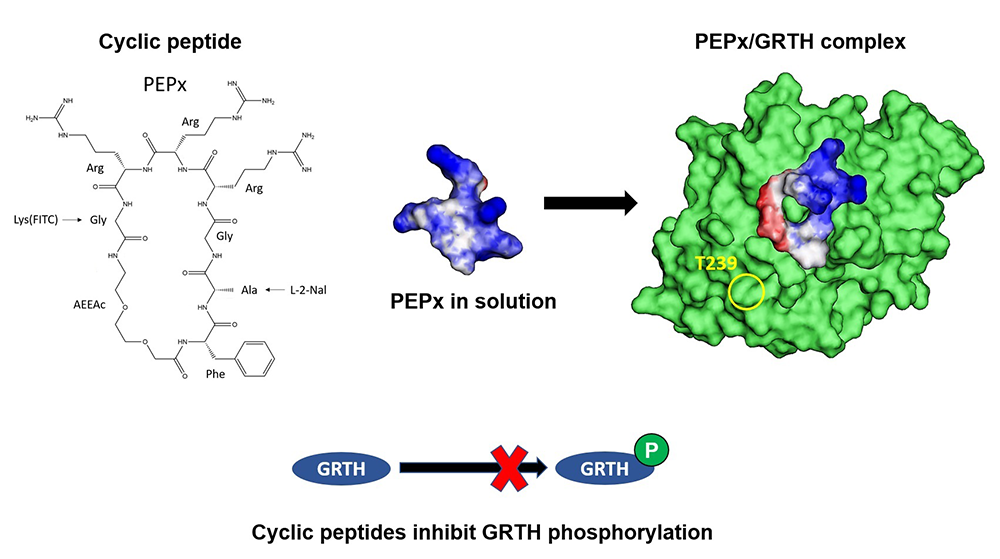
During the past year, we determined that cyclic peptides that fit the shallow pocket of the GRTH/DDX5/PKA interphase are preferred compounds to block GRTH phosphorylation and amenable for use in the development of an oral male contraceptive [Reference 4] (Figure 1). In this regard, these cyclic peptides (PEP0, PEP1, and PEP2) have been designed and synthesized as promising therapeutic agents. PEP1 and PEP2, revealed by fluorescein isothiocyanate (FITC), showed effective internalization in COS-1 cells and seminiferous tubules after a 4-hour treatment. We observed a dose-dependent inhibitory effect on GRTH phosphorylation in a COS-1 stable cell line expressing GRTH, with significant reduction in pGRTH protein observed after 8–16 hour treatments. CETSA (cellular thermal shift assay) showed compound binding resulting in thermal stabilization of the soluble non–pGRTH protein when compared with control peptides. Increased efficiency in a fluorescence resonance energy transfer (FRET) assay revealed the interaction of the cyclic peptide with GRTH. Exposure of a culture of seminiferous tubules to these compounds resulted in significant inhibition of the pGRTH protein species. Similar results were obtained with the compound that lacks FITC. Taken together, effective internalization and targeted reduction in the expression of pGRTH by cyclic peptides provide a promising angle to develop effective compounds for use as a non-hormonal male contraceptive.
Figure 1. Cyclic peptides inhibit GRTH phosphorylation.
Peptide x (left) (1FAGRRRG7) represents one of the simplest cyclic peptides that accommodates stringent conditions and the main elements of the pharmacophore. The molecular scaffold provided the basis for the design of cyclic peptides (CPs) that bind to GRTH. A computational multistage cyclization procedure predicted the binding modes at the interphase. The simulations indicated that, in solution, the CPs are quite flexible, an attribute that permits adaptation to the GRTH/PKA interphase (center, right) to exert its inhibitory function on the phosphorylation of GRTH at T239 (yellow circle, right). Binding of the CPs to GRTH was confirmed by thermal stabilization of non-phospho–GRTH and increased efficiency in FRET assay [Reference 4].
Also, work in transcriptome profiles of germ cell isolated form GRTH–KI mice vs. those of wild type (WT) mice, using the RNA-Seq technique, has provided further insights linking phospho-GRTH to histone ubiquitination and acetylation essential for chromatin compaction and spermatid elongation during spermiogenesis [Reference 5].
We also initiated studies on the role of phosphorylated GRTH in the storage of messages in the CB. We observed absence of pGRTH from the CB of GRTH KI mice (with an insertion of the R242H human mutation that abolishes GRTH phosphorylation at T239 and spermatogenesis). RNA-Seq analysis of mRNA isolated from the CB revealed that the abundance of 947 genes was reduced and that of 474 genes increased [Reference 5]. Transcripts related to spermatid development, differentiation, and chromatin remodeling were reduced; in contrast those encoding factors involved in RNA transport, regulation, surveillance, and transcriptional and translational regulation were increased in the CB of KI mice, which was validated by qPCR. Transcripts of several initiation factors (eIF4e, 4ebp2, 3land 3m), together with mRNAs related to the 60S subunit (RpL101/RPlp0), were increased and accumulated in CB, mRNAs that could not be transported from the CB to polyribosomes for translation; instead, they remain stored in the CB in KI mice owing to loss of pGRTH [Reference 6]. Our studies demonstrated the importance of phospho–GRTH in the maintenance of the structure of the CB and its role in the storage and stability of germ cell–specific mRNAs during spermiogenesis.
Publications
- Dufau ML, Kavarthapu R. Gonadotropin regulated testicular RNA helicase, two decades of studies on its structure function and regulation from its discovery opens a window for development of a non-hormonal oral male contraceptive. Front Endocrinol (Lausanne) 2019;10:576–586.
- Kavarthapu R, Anbazhagan R, Raju M, Morris CT, Pickel J, Dufau ML. Targeted knock-in mice with a human mutation in GRTH/DDX25 reveals the essential role of phosphorylated GRTH in spermatid development during spermatogenesis. Hum Mol Genet 2019;28:2561–2572.
- Raju M, Hassan SA, Kavarthapu R, Anbazhagan R, Dufau ML. Characterization of the phosphorylation site of GRTH/DDX25 and protein kinase A binding interface provides structural basis for the design of a non-hormonal male contraceptive. Sci Rep 2019;9:42857–42867.
- Raju M, Kavarthapu R, Anbazhagan R, Hassan SA, Dufau ML. Blockade of GRTH/DDX25 phosphorylation by cyclic peptides provides an avenue for developing a non-hormonal contraceptive. J Med Chem 2021;64:14715–14727.
- Kavarthapu R, Anbazhagan R, Sharma AK, Shiloach J, Dufau ML. Linking phospho-gonadotropin regulated testicular RNA helicase (GRTH/DDX25) to histone ubiquitination and acetylation essential for spermatid development during spermiogenesis. Front Cell Dev Biol 2020;8:310–321.
- Anbazhagan R, Kavarthapu R, Coon SL, Dufau ML. Role of phosphorylated-gonadotropin regulated testicular RNA helicase (GRTH/DDX25) in the regulation of cell specific mRNAs in chromatoid bodies during spermatogenesis. Front Cell Dev Biol 2020;8:580019.
Collaborators
- Sergio A. Hassan, PhD, Bioinformatics and Computational Biosciences Branch, NIAID, Bethesda, MD
Contact
For more information, email dufau@helix.nih.gov or visit https://irp.nih.gov/pi/maria-dufau.