Hippocampal Interneurons and Their Role in the Control of Network Excitability

- Chris J. McBain, PhD, Head, Section on Cellular and Synaptic Physiology
- Ramesh Chittajulla, PhD, Staff Scientist
- Kenneth Pelkey, PhD, Staff Scientist
- James D'Amour, PhD, Postdoctoral Fellow
- June Hoan Kim, PhD, Postdoctoral Fellow
- Vivek Madahavan, PhD, Postdoctoral Fellow
- Geoffrey Vargish, PhD, Postdoctoral Fellow
- Steven Hunt, Biologist
- Xiaoqing Yuan, MSc, Biologist
- Tyler Ekins, BSc, Predoctoral Fellow
- Connie Mackenzie-Gray Scott, Bsc, Predoctoral Fellow
- Areg Peltekian, BSc, Postbaccalaureate Fellow
- Anna Vlachos, BSc, Postbaccalaureate Fellow
Cortical and hippocampal GABAergic inhibitory interneurons (INs) are ‘tailor-made’ to control cellular and network excitability by providing synaptic and extrasynaptic input to their downstream targets via GABAA and GABAB receptors. The axons of this diverse cell population make local, short-range projections (although some subpopulations project their axons over considerable distances) and release the inhibitory neurotransmitter gamma-aminobutyric acid (GABA) onto a variety of targets. Distinct cohorts of INs regulate sub- and supra-threshold intrinsic conductances, regulate Na+- and Ca2+-dependent action-potential generation, modulate synaptic transmission and plasticity, and pace both local- and long-range large-scale synchronous oscillatory activity. An increasing appreciation of the roles played by INs in several neural-circuit disorders, such as epilepsy, stroke, Alzheimer’s disease, and schizophrenia, has seen this important cell type take center stage in cortical circuit research. With almost 30 years of interest in this cell type, the main objectives of the lab have been to understand: (1) the developmental trajectories taken by specific cohorts of INs as they populate the nascent hippocampus and cortex; (2) how ionic and synaptic mechanisms regulate the activity of both local-circuit GABAergic INs and principal neurons (PN) at the level of small, well defined networks; and (3) how perturbations in their function alter the cortical network in several neural circuit disorders. To this end, we use a variety of electrophysiological, imaging, optogenetic, immunohistochemical, biochemical, molecular, and genetic approaches with both wild-type and transgenic animals.
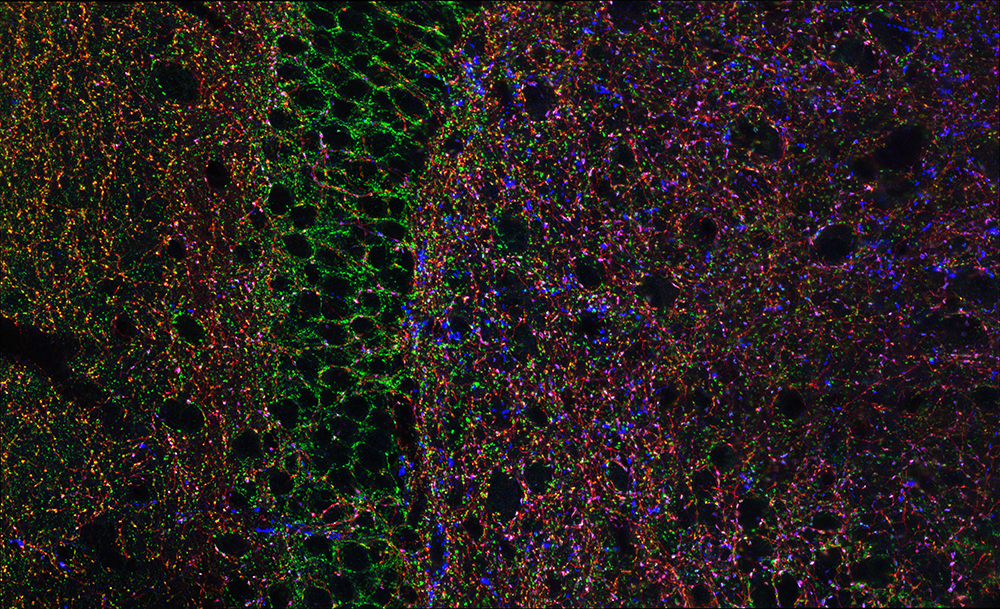
Figure 1. Hippocampal GABAergic terminals
Three major types of GABAergic terminal are shown in the hippocampal dentate gyrus (red, blue, and green punch). The black negative space shows the cell bodies of principal cells innervated by the GABAergic terminals.
Aberrant sorting of hippocampal complex pyramidal cells in type I lissencephaly alters topological innervation.
Layering has been a long-appreciated feature of higher-order mammalian brain structures, but the extent to which it plays an instructive role in synaptic specification remains unknown. We examined the formation of synaptic circuitry under cellular heterotopia (which occurs when neurons do not migrate properly during the early development of the fetal brain) in the hippocampal CA1 region, using a mouse model of the human neuro-developmental disorder type I lissencephaly. We identified calbindin-expressing principal cells that are mis-positioned under cellular heterotopia. Ectopic calbindin-expressing principal cells develop relatively normal morphological features but stunted intrinsic physiological features. Regarding network development, a connectivity preference for cholecystokinin-expressing interneurons to target calbindin expressing principal cells is diminished. Moreover, in vitro gamma oscillatory activity is less synchronous across heterotopic bands, and mutants are less responsive to pharmacological antagonism of cholecystokinin-containing interneurons. With this study, we hope to aid not only in our understanding of how cellular networks form but highlight vulnerable cellular circuit motifs that might be generalized across disease states.
Emergence of non-canonical parvalbumin-containing interneurons in hippocampus of a murine model of type I lissencephaly
Type I lissencephaly is a neuronal migration disorder caused by haploinsuffiency of the LIS1 gene and is characterized in humans by agyria, mis-lamination of brain structures, developmental delays, and epilepsy. We investigated the impact of the LIS1 mutation on the cellular migration, morphophysiology, microcircuitry, and genomics of mouse hippocampal CA1 parvalbumin-containing inhibitory interneurons (PV+INTs). We found that wild-type (WT) PV+INTs consist of two physiological subtypes (80% fast-spiking [FS], 20% non-fast-spiking [NFS]) and four morphological subtypes (basket, axo-axonic, bistratified, radiatum-targeting). We also discovered that cell-autonomous mutations within interneurons disrupt morphological development of PV+INTs, which results in the emergence of a non-canonical “intermediate spiking (IS)” subset of PV+INTs. In the GlobalLis mutant mouse (a mouse model of lissencephaly), IS/NFS cells become the dominant PV+INT subtypes (56%) and the percentage of FS cells shrinks to 44%. We also found that IS/NFS cells are prone to entering depolarizing block, causing them to temporarily lose the ability to initiate action potentials and control network excitation, potentially promoting seizures. Also, single-cell nuclear RNA-Seq of PV+INTs revealed several mis-regulated genes related to morphogenesis, cellular excitability, and synapse formation.
NMDAR–mediated transcriptional control of gene expression in the specification of interneuron subtype identity.
Medial ganglionic eminence (MGE)–derived parvalbumin (PV)+, somatostatin (SST)+, and neurogliaform (NGFC)–type cortical, and hippocampal interneurons have distinct molecular, anatomical, and physiological properties. However, the molecular mechanisms regulating their diversity remain poorly understood. Using a single-cell transcriptomics, we showed that the obligate NMDA–type glutamate receptor (NMDAR) subunit gene Grin1 mediates subtype-specific transcriptional regulation of gene expression in MGE–derived interneurons, leading to altered subtype identities. Notably, MGE–specific conditional Grin1 loss results in a systemic downregulation of diverse transcriptional, synaptogenic, and membrane excitability regulatory programs. Such widespread gene-expression abnormalities mirror aberrations that are typically associated with neuro-developmental disorders, particularly schizophrenia. Our study thus provides a road map for the systematic examination of NMDAR signaling in interneuron subtypes, revealing potential MGE–specific genetic targets that could instruct future therapies for psychiatric disorders.
Intrinsic electrophysiological properties predict variability in morphology and connectivity among striatal parvalbumin-expressing Pthlh cells.
Determining the cellular content of the nervous system in terms of cell types and the rules of their connectivity represents a fundamental challenge to the neurosciences. The recent advent of high-throughput techniques, such as single-cell RNA-Seq, has allowed for greater resolution in the identification of cell types and/or states. Although most of the current neuronal classification schemes comprise discrete clusters, several recent studies have suggested that, perhaps especially within the striatum, neuronal populations exist in continua with regard to both their molecular and electrophysiological properties. Whether these continua are stable properties, established during development, or whether they reflect acute differences in activity-dependent regulation of critical genes is currently unknown. We set out to determine whether gradient-like molecular differences in the recently described Pthlh–expressing inhibitory interneuron population, which contains the Pvalb–expressing cells, correlate with differences in morphological and connectivity properties. We showed that morphology and long-range inputs correlate with a spatially organized molecular and electrophysiological gradient of Pthlh interneurons, suggesting that the processing of different types of information (by distinct anatomical striatal regions) has different computational requirements.
Additional Funding
- James D’Amour was funded by an NIGMS PRAT Fellowship.
Publications
- D’Amour JA, Ekins TG, Ghanatra S, Yuan X, McBain CJ. Aberrant sorting of hippocampal complex pyramidal cells in Type I Lissencephaly alters topological innervation. eLife 2020;9:e55173.
- Ekins TG, Mahadevan V, Zhang Y, Akgul G, Petros T, McBain CJ. Emergence of non-canonical parvalbumin-containing interneurons in hippocampus of a murine model of Type I lissencephaly. eLife 2020;9:e62373.
- Vormstein-Schneider DC, Lin JD, Pelkey KA, Chittajallu R, Guo B, Arias Garcia M, Allaway K, Sakopoulos S, Schneider G, Stevenson O, Vergara J, Sharma J, Zhang Q, Franken TP, Smith J, Ibrahim LA, Mastro K, Sabri E, Huang S, Favuzzi E, Burbridge T, Xu Q, Guo L, Vogel I, Sanchez V, Saldi GA, Yuan X, Zaghloul KA, Devinsky O, Sabatini B, Batista-Brito R, Reynolds J, Feng G, Fu Z, McBain CJ, Fishell GJ, Dimidschstein J. Viral manipulation of functionally distinct neurons from mice to humans. Nat Neurosci 2020;23:1629–1636.
- Bengtsson Gonzales C, Hunt S, Munoz Manchado A, McBain CJ, Hjerling-Leffler J. Intrinsic electrophysiological properties predict variability in morphology and connectivity among striatal Parvalbumin-expressing Pthlh-cells. Sci Rep 2020;10:15620.
- Mahadevan V, Mitra A, Zhang Y, Peltekian A, Chittajallu R, Esnault C, Maric D, Rhodes C, Pelkey KA, Dale R, Petros T, McBain CJ. NMDAR-mediated transcriptional control of gene expression in the specification of interneuron subtype identity. Front Mol Neurosci, Mol Signal Pathways 2021;10:3389.
Collaborators
- Jordan Dimidschstein, PhD, The Broad Institute, Cambridge, MA
- Gordon Fishell, PhD, The Broad Institute, Cambridge, MA
- Timothy J. Petros, PhD, Unit on Cellular and Molecular Neurodevelopment, NICHD, Bethesda, MD
- Kareem Zaghloul, MD, PhD, Functional and Restorative Neurosurgery Unit, NINDS, Bethesda MD
Contact
For more information, email mcbainc@mail.nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/mcbain or https://dir.ninds.nih.gov/Faculty/Profile/chris-mcbain.html.