The Molecular Mechanics of Eukaryotic Translation Initiation

- Jon Lorsch, PhD, Chief, Laboratory on the Mechanism and Regulation of Protein Synthesis
- Jagpreet Nanda, PhD, Staff Scientist
- Fujun Zhou, PhD, Research Fellow
The goal of our research is to elucidate the molecular mechanisms underlying the initiation phase of protein synthesis in eukaryotic organisms. We use the yeast Saccharomyces cerevisiae as a model system and employ a range of approaches, from genetics to biochemistry to structural biology, in collaboration with Alan Hinnebusch's and Tom Dever’s labs and several other research groups around the world.
Eukaryotic translation initiation is a key control point in the regulation of gene expression. It begins when an initiator methionyl tRNA (Met-tRNAi) is loaded onto the small (40S) ribosomal subunit. Met-tRNAi binds to the 40S subunit as a ternary complex (TC) with the GTP–bound form of the initiation factor eIF2. Three other factors, eIF1, eIF1A, and eIF3, also bind to the 40S subunit and promote the loading of the TC. The resulting 43S preinitiation complex (PIC) is then loaded onto the 5′ end of an mRNA with the aid of eIF3 and the eIF4 group of factors: the RNA helicase eIF4A; the 5′ 7-methylguanosine cap-binding protein eIF4E; the scaffolding protein eIF4G; and the 40S subunit– and RNA–binding protein eIF4B. Both eIF4A and eIF4E bind to eIF4G and form the eIF4F complex. Once loaded onto the mRNA, the 43S PIC is thought to scan the mRNA in search of an AUG start codon. The process is ATP–dependent and likely requires several RNA helicases, including the DEAD–box protein Ded1p. Recognition of the start site begins with base pairing between the anticodon of tRNAi and the AUG codon. Base pairing then triggers downstream events that commit the PIC to continuing initiation from that point on the mRNA, events that include ejection of eIF1 from its binding site on the 40S subunit, movement of the C-terminal tail (CTT) of eIF1A, and release of phosphate from eIF2, which converts eIF2 to its GDP–bound state. In addition, the initiator tRNA moves from a position that is not fully engaged in the ribosomal P site [termed P(OUT)] to one that is [P(IN)], and the PIC as a whole converts from an open conformation, which is conducive to scanning to a closed one, which is not. At this stage, eIF2•GDP dissociates from the PIC, and eIF1A and a second GTPase factor, eIF5B, coordinate joining of the large ribosomal subunit to form the 80S initiation complex. In a process that appears to result in conformational reorganization of the complex, eIF5B hydrolyzes GTP and then dissociates along with eIF1A.
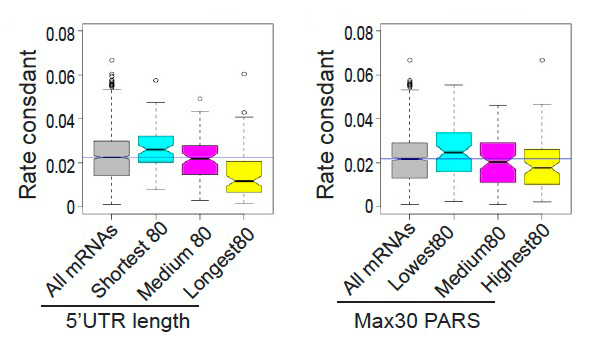
In 2021, we continued to make progress on implementation of the RecSeq platform that we had developed to allow measurement of translation initiation rates transcriptome-wide in a fully reconstituted in vitro system. We used the system to measure the rate constants for recruitment of most yeast mRNAs, something that has never been done before. Our data indicate that the length of the 5′-untranslated region (5′-UTR) and the amount of structure in the 5′-UTR both inversely correlate with the rate of mRNA recruitment. We also compiled a large amount of data about the role of the RNA helicase initiation factor Ded1 in the mRNA recruitment process using this system. Ded1 preferentially accelerates the rate of recruitment of mRNAs with long, structured 5′-UTRs, consistent with data from in vivo experiments from Alan Hinnebusch's lab. We expanded these studies to probe the roles of the factor eIF4A and test the effects of competition among mRNAs for limiting 40S ribosomal subunits in the system. Such studies are significant because of debates in the field around whether certain pathologies (e.g., ribosomopathies) affect gene expression by altering translation of certain mRNAs as a result of differential competition among mRNAs that are inherently well or poorly translated, or because ribosomes lacking or containing different components (e.g., ribosomal proteins) have different inherent specificities for certain mRNAs owing to ribosome-mRNA interactions ("specialized ribosomes"). The studies are breaking new ground by allowing us to actually measure rates of mRNA recruitment to 40S ribosomes transcriptome-wide under defined conditions in a system in which we can isolate translation initiation from other cellular processes, such as transcription and mRNA decay.
The pandemic slowed progress by our group, and a senior member of the lab (Jagpreet Nanda) departed to take a position as a Scientific Review Officer at NICHD, but despite these challenges we have progressed significantly. We have now hired a new member of the lab, Meizhen Hou, who has exceptional experience in protein expression and purification and will thus be a great asset to the lab and the NICHD IRP. We also expect to have a postbaccalaureate student joining the lab this summer, after her graduation from Wellesley College. This will bring staffing in the lab to two biologists and a postbaccalaureate.
Figure 1.
Figure showing the correlation between rate constants for mRNA recruitment, measured using the RecSeq platform and 5′-UTR length (left) or structure (right). PARS is a measure of RNA structure.
Additional Funding
- Funding is provided via a Memorandum of Understanding (MOU) between NIGMS and NICHD.
Publication
- Gulay S, Gupta N, Lorsch JR, Hinnebusch AG. Distinct interactions of eIF4A and eIF4E with RNA helicase Ded1 stimulate translation in vivo. eLife 2020;9:e58243.
Collaborators
- Thomas Dever, PhD, Section on Protein Biosynthesis, NICHD, Bethesda, MD
- Alan Hinnebusch, PhD, Section on Nutrient Control of Gene Expression, NICHD, Bethesda, MD
- Venkatraman Ramakrishnan, PhD, MRC Laboratory of Molecular Biology, Cambridge, United Kingdom
Contact
For more information, email jon.lorsch@nih.gov or visit https://irp.nih.gov/pi/jon-lorsch.