Signaling Interpretation during Vertebrate Embryogenesis

- Katherine W. Rogers, PhD, Head, Unit on Developmental Signaling
- William K. Anderson, BS, Research Technician
- Catherine E. Rogers, BS, Postbaccalaureate Fellow
Our lab seeks to understand cell-lineage differentiation, gene regulation, and how non-coding DNA elements and the 3D architecture of chromosomes contribute to such processes during development and disease. We are also interested in early mammalian development as a system in which to decipher how cells make lineage decisions and how gene-regulatory networks are established.
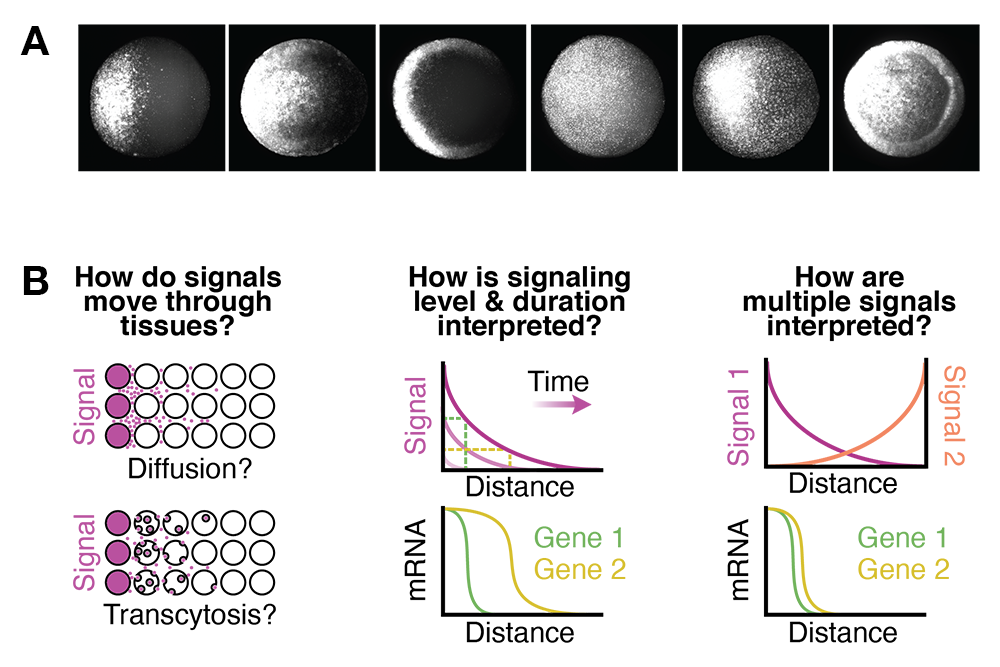
To create the different tissues required in healthy adults, embryos must activate fate-specifying genes in a variety of stereotyped positions (Figure 1A), which is mediated by signaling molecules that spread though embryonic tissues. It is thought that signaling levels and dynamics regulate gene expression, and that position-specific signaling differences underlie the diverse gene expression patterns required for normal development. However, it is not clear what signaling features are 'read out' by genes and how those features are converted into differential gene expression during vertebrate embryogenesis.
We investigate how signaling molecules spread through embryonic tissues, how signaling levels and dynamics are decoded, and how many pathways cooperate to pattern the body plan (Figure 1B). To directly examine these processes, we need precise in vivo experimental manipulations that are difficult to achieve with conventional genetic and pharmacological methods. In contrast, molecular optogenetic approaches can offer tunable, reversible manipulation of biological processes with excellent temporal (seconds) and spatial (subcellular) resolution, and such approaches have been robustly demonstrated both in vitro and in vivo. Using the microscopy-friendly zebrafish embryo as a vertebrate model system, our lab will harness established optogenetic approaches and develop new ones to investigate patterning mediated by conserved signaling pathways.
Figure 1.
A. Gene expression in the zebrafish embryo
B. outstanding developmental questions
How do signaling molecules move through tissues?
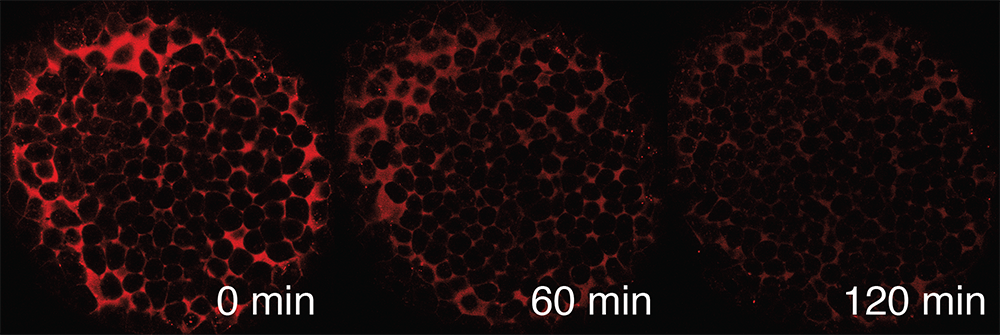
The distribution of signaling molecules within developing tissues helps determine patterns of gene expression. Competing models have been proposed to explain how signaling-molecule distributions are established: signals may diffuse away from producing cells through the extracellular space, move through cells (trans-cytosis), or be confined to the producing cells themselves. We will develop optogenetic tools to probe how factors, including extracellular diffusion and trans-cytosis, affect signaling molecule distribution and use these with in vivo methods, including FRAP (fluorescence recovery after photobleaching) and FDAP (fluorescence decay after photoactivation) (Figure 2), to directly measure signaling molecule mobility and stability. This will help determine how signaling-molecule distribution is regulated during zebrafish embryogenesis.
What information is encoded in signaling gradients?
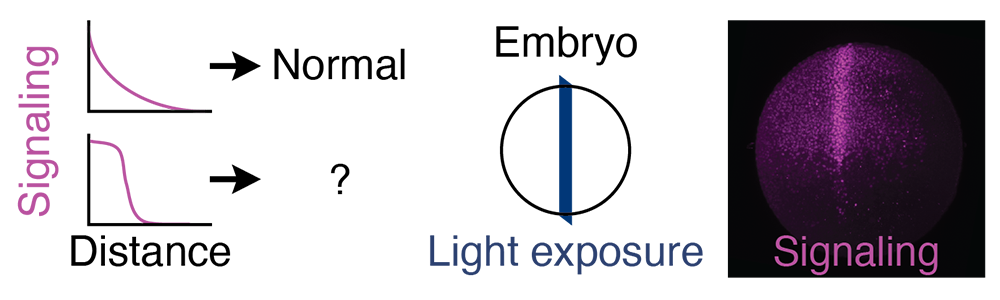
Signaling gradients are found in developing tissues from the fly wing precursor to the mammalian neural tube. The classic morphogen model proposes that the precisely graded distribution of signaling is important because genes are activated by different signaling levels. Alternatively, a simple signaling asymmetry may suffice to pattern tissues in some contexts. The relatively subtle signaling perturbations required to distinguish between these models can be difficult to achieve in vivo. We will develop optogenetic approaches and use them with a digital micromirror device to introduce novel signaling distributions in zebrafish embryos and assess patterning consequences (Figure 3). This will determine the spatiotemporal signaling requirements for normal tissue patterning during early zebrafish development.
How are signaling levels, dynamics, and combinations interpreted in the embryo?
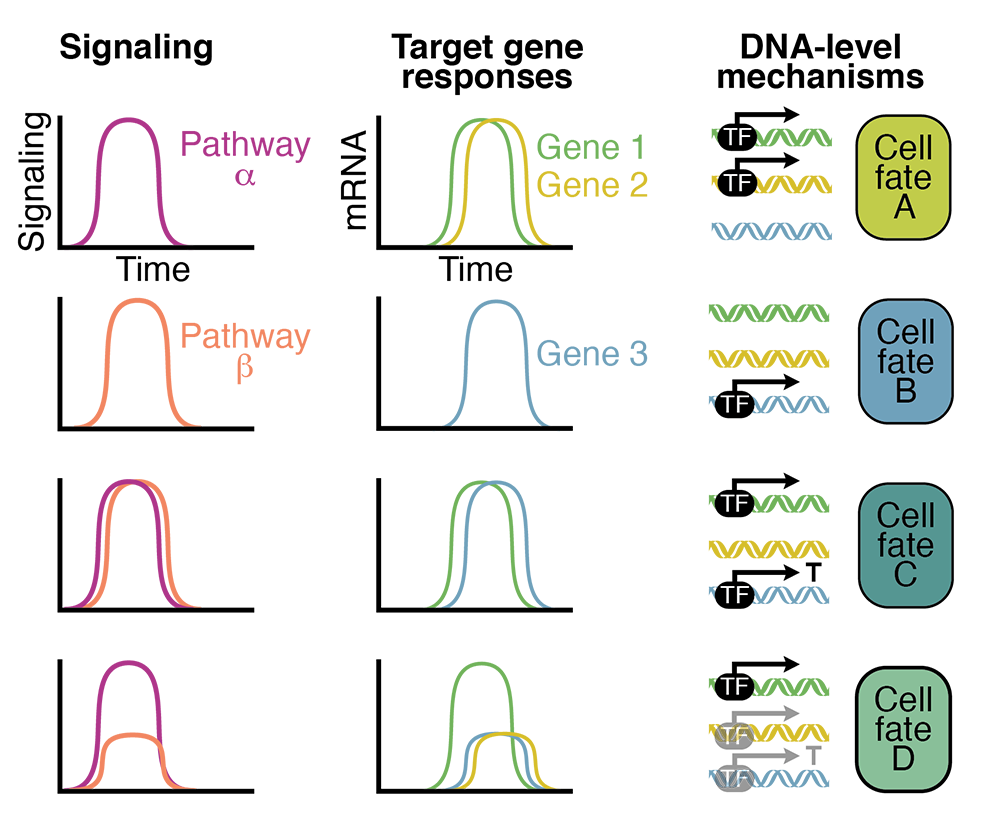
Cells in developing tissues experience a variety of signaling levels and dynamics, as well as simultaneous signaling from several pathways. How do different genes respond to these inputs? We seek to determine the input/output relationship between signaling and gene expression during early vertebrate embryogenesis. To achieve this, we will develop orthogonal optogenetic tools to manipulate signaling levels and dynamics in zebrafish embryos. We will characterize gene responses and investigate the DNA–level mechanisms responsible for differential responses (Figure 4). This will help elucidate which features of signaling encode information and help explain how the diverse gene expression patterns needed to produce healthy adults are robustly generated.
Publications
- Rogers KW, ElGamacy M, Jordan BM, Müller P. Optogenetic investigation of BMP target gene expression diversity. eLife 2020;e58641.
- Rogers KW, Müller P. Optogenetic approaches to investigate spatiotemporal signaling during development. In: Gradients and Tissue Patterning. Small S, Briscoe J, eds, 2020;37–77.
- Rogers KW, Lord ND, Gagnon JA, Pauli A, Zimmerman S, Aksel DC, Reyon D, Tsai SQ, Joung JK, Schier AF. Nodal patterning without Lefty inhibitory feedback is functional but fragile. eLife 2017;e28785.
- Pomreinke AP, Soh GH, Rogers KW, Bergmann JK, Bläßle AJ, Müller P. Dynamics of BMP signaling and distribution during zebrafish dorsal-ventral patterning. eLife 2017;e25861.
Contact
For more information, email katherine.rogers@nih.gov or visit https://www.nichd.nih.gov/research/atNICHD/Investigators/rogers.