The Biophysics of Protein-Lipid Interactions in Influenza and Coronavirus, Malaria, and Muscular Dystrophy

- Joshua Zimmerberg, MD, PhD, Head, Section on Integrative Biophysics
- Paul S. Blank, PhD, Staff Scientist
- Svetlana Glushakova, MD, PhD, Staff Scientist
- Matthias Garten, PhD, Visiting Fellow
- Irene Jimenez Munguia, PhD, Visiting Fellow
- Yuto Kegawa, PhD, Visiting Fellow
- Hang Waters, MS, Biologist
- Jennifer Petersen, PhD, Electron Microscopist
- Elena Mekhedov, MA, Contractor
- Tatyana I. Tenkova-Heuser, PhD, Contractor
- Ludmila Bezrukov, MS, Chemist
- John E. Heuser, MD, Senior Biophysicist
- Glen Humphrey, PhD, Guest Researcher
- Adriana Golding, PhD, Postdoctoral Intramural Research Training Award Fellow
- Blaise A. Stearns, BS, Postbaccalaureate Fellow
- Garrett Tisdale, BA, Postbaccalaureate Trainee
Eukaryotic life must create the many shapes and sizes of the system of internal membranes and organelles that inhabit the variety of cells in nature, membranes that must remodel for cells to repair damaged plasmalemma and deal with infectious agents such as viruses and parasites. Such basic membrane mechanisms must be highly regulated and highly organized in various hierarchies in space and time to allow the organism to thrive despite environmental challenges, genetic instability, unpredictable food supply, and physical trauma. We are using our expertise and the techniques we perfected over the years to address various biological problems that have in common the underlying regulation or disturbance of protein/lipid interactions. The overall goal of this project is to determine the physico-chemical mechanisms of membrane remodeling in cells.
Identification of inhibitors of coronaviral entry
Some of the most dangerous human and animal pathogens, in particular coronavirus, influenza virus, and human immunodeficiency virus (HIV), are enveloped viruses. For these viruses, receptor binding and entry are accomplished by a single viral envelope protein (termed the spike or fusion protein), the structural changes in which trigger the remodeling and merger of the viral and target cellular membranes. We are focusing our continuing research on the physiology of deadly viruses and parasites towards fighting the COVID-19 pandemic, in areas closest to our expertise: helping to produce drugs against COVID-19 and producing viral-like particles of SARS-CoV-2. While some coronaviruses are endemic and cause only a common cold, others, those like SARS-CoV-2, are highly pathogenic and cause COVID-19. Previously the coronavirus strain SARS-CoV caused a severe acute respiratory syndrome outbreak in Asia in 2003, and during 2013 the Middle East respiratory syndrome (MERS) emerged with clinical symptoms similar to SARS, and the causative agent was named MERS-CoV. Many clinical trials are under way in search of treatments for COVID-19. Anti-inflammatory drugs such as corticosteroids are being used, and some treatments have received emergency use authorization by the FDA, such as monoclonal antibody therapy and remdesivir. However, more effective therapies are needed to combat the COVID-19 pandemic.
To combat COVID-19, high throughput screening (HTS) of thousands of candidate compounds that may block or reduce SARS-CoV-2 infection of target tissues is a powerful tool in drug discovery. However, the SARS-CoV-2 pathogen requires a biological safety level 3 (BSL-3), whereas most HTS facilities are only BSL-2. To overcome this obstacle and meet the desperate need for HTS of compounds with SARS-CoV-2 antiviral activity, an HTS screening assay was developed using pseudotyped particles (PP). PP serve as mimics for viral binding and entry into target cells, are BSL-2–compatible, and can be used for HTS. PP contain viral-envelope proteins such as the spike protein (S protein), which mediates viral entry into target cells, but carry a fluorescence-based reporter gene instead of the viral genome, and thus display the necessary viral coat proteins for host-receptor and membrane interactions without the capacity for replication. Similar PP–based BSL-2 viral entry assays have been successfully applied to HTS campaigns against the Ebola, influenza, and human immunodeficiency viruses (HIV). In a collaboration with scientists at NCATS, we used negative-stain electron microscopy and immunogold labeling to evaluate the structure of S proteins and quantify their distribution on PP that were used in their HTS assays for compounds with anti-SARS-CoV-2 activity. The technique was used countless times over this year to continuously monitor quality control for the PP for various SARS-CoV-2 S protein variants, as well as control PP. The PP were created by co-transfection of a plasmid of the S protein of a coronavirus and a plasmid for the Gag protein of a murine leukemia virus. For measurement of S–mediated entry, luciferase was included in the PP; successful target-cell entry was measured as luciferase activity. In this way, our colleagues identified inhibitors of S–mediated cell entry in a high throughput screen of an approved drugs library with SARS-S and MERS-S pseudotyped particle entry assays. Six compounds were discovered (cepharanthine, abemaciclib, osimertinib, trimipramine, colforsin, and ingenol) to be broad-spectrum inhibitors of S–mediated entry. The work could contribute to the development of effective treatments against the initial stage of viral infection and provide mechanistic information that might aid in the design of new drug combinations for clinical trials for COVID-19 patients. After further testing in a SARS-CoV-2 live-virus cytopathic-effect (CPE) assay and removing cytotoxic compounds, six out of seven entry inhibitors were able to rescue the CPE of SARS-CoV-2 infection, indicating the utility of these PP entry assays.
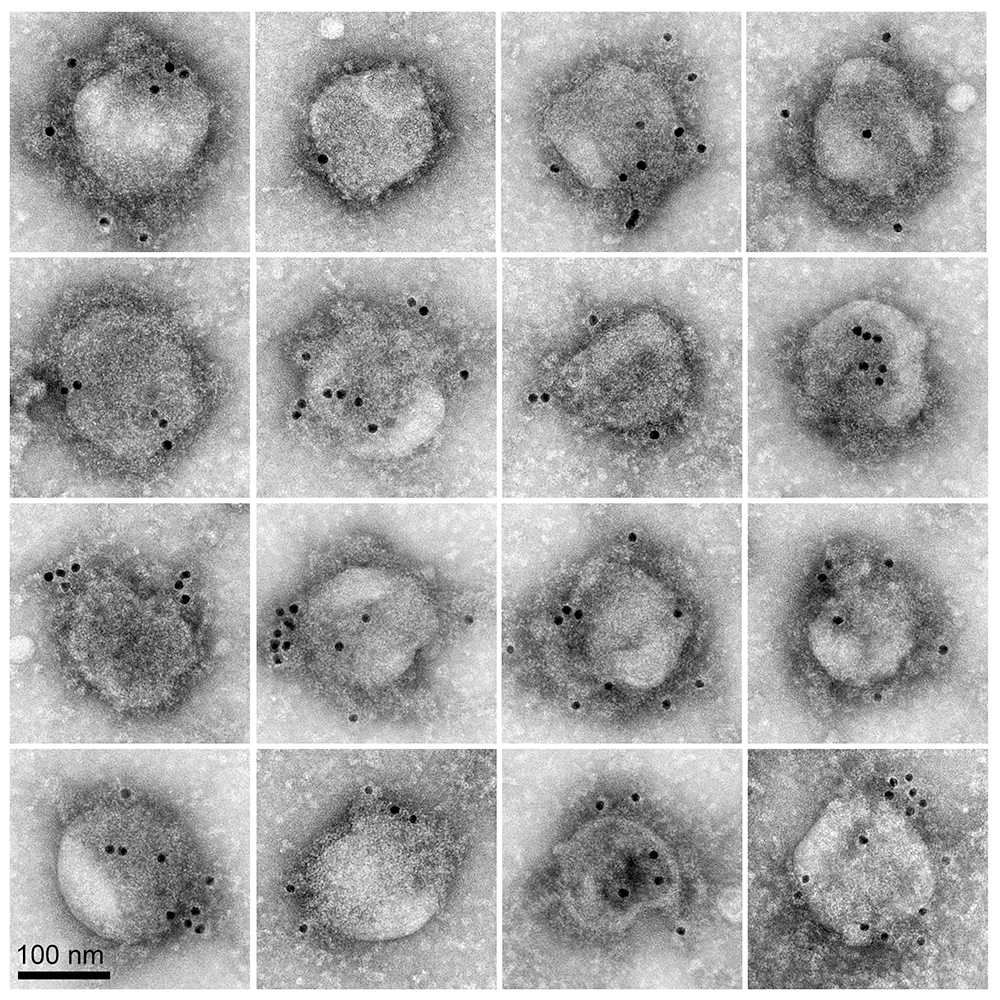
Figure 1. SARS-CoV-2 virus entry particles
Negative stain microscopy image shows a montage of “SARS-CoV-2 viral entry mimics,” or pseudotyped particles (PPs). The PPs are membrane-encased, virus-sized spheres decorated with the SARS-CoV-2 spike protein. PPs are non-infectious, so can be safely used to study how the spike protein mediates binding to and entry into host cells and to identify compounds that block spike-mediated viral entry in high-throughput drug screening assays. The spike proteins appear as a dense fuzz surrounding the 150 nm–diameter particles and are labeled with 10-nm immunogold (black dots).
New assays for enveloped viral fusion
Survival of the SARS-COV-2 virus particle for person-to-person transmission and its ability to fuse with the host cell membrane are highly dependent on the lipid composition of the viral envelope. Determining the lipid composition and physical properties of attenuated virus and virus-like particles (VLPs) will permit the study of membrane stability and fusogenicity under physiological conditions. Our goal is to give the scientific community a VLP to efficiently study virus infection in widely available non-BL2 laboratory space and yield enough lipid for analysis. Viral fusion is a critical step in the entry pathway of enveloped viruses and remains a viable target for antiviral exploration. The current approaches to studying fusion mechanisms include ensemble fusion assays, high-resolution cryo-transmission electron microscopy (TEM), and single-molecule fluorescence-based methods. While these methods have provided invaluable insights into the dynamic events underlying fusion processes, they come with their own limitations, which often include extensive data and image analysis in addition to experimental time and technical requirements. Our work proposes the use of the spin-spin T2 relaxation technique as a sensitive bioanalytical method for the rapid quantification of interactions between viral fusion proteins and lipids in real time. In the study, new liposome-coated iron oxide nanosensors (LIONs), which mimic as magnetic-labeled host membranes, are reported to detect minute interactions between the membrane and hemagglutinin (HA), influenza's fusion glycoprotein. The influenza fusion protein's interaction with the LION membrane is detected by measuring changes in the sensitive spin-spin T2 magnetic relaxation time using a bench-top nuclear-magnetic resonance (NMR) instrument. More data is gleaned from including the fluorescent dye DiI into the LION membrane. In addition, the effects of environmental factors on protein-lipid interaction that affect fusion, such as pH, time of incubation, trypsin, and cholesterol, were also examined. Furthermore, using VLPs we demonstrated the efficacy and sensitivity of the spin-spin T2 relaxation assay in quantifying similar protein/lipid interactions with more native configurations of HA. Shorter domains derived from HA were used to start a reductionist path to identify the parts of HA responsible for the NMR changes observed. Also, the known fusion inhibitor Arbidol was employed in our spin-spin T2 relaxation-based fusion assay to demonstrate the application of LIONs in real-time monitoring of this aspect of fusion for evaluation of potential fusion inhibitors.
Mechanisms of membrane electro-poration
Living cells are open non-equilibrium systems. To exist, a cell requires precisely controlled maintenance of gradients in the chemical potential between the extracellular environment, the cytoplasm, and the lumen of organelles, of many constituents. The amphiphilic nature of lipid molecules, self-assembling into lipid bilayers, provides an extremely low permeability barrier to both electrolytes and large non-electrolytes. For the processes of life, a continuous exchange of matter must occur across all membranes. For example, uptake of large molecules and compounds from the outside occurs via endocytosis, phagocytosis, and macro- and micro-pinocytosis, secretion occurs via exocytosis, and intracellular protein trafficking via transport vesicles between endoplasmic reticulum, Golgi apparatus, endosomes, and lysosomes. The movement of such membrane-bound cargo dictates membrane recycling, or cells and organelles would be incapable of maintaining their volumes and shapes. These ubiquitous and multifarious events, plus the accommodation of lipid bilayers to membrane proteins and transient pores, all require the lipid bilayer to change its topology. Physical models of such topological changes, essential to life, must take into account the energy of membrane deformations within the framework of an adequate theory of elasticity. Although transport of molecules into cells via electro-poration is a common biomedical procedure, its protocols are often based on trial and error. Despite a long history of theoretical effort, the underlying mechanisms of cell-membrane electro-poration have not been sufficiently elucidated, in part, because many independent fitting parameters need to link theory with experiment.
In this project, we investigate whether the electro-poration behavior of a reduced cell membrane is consistent with time-resolved, atomistic, molecular dynamics (MD) simulations of phospholipid bilayers responding to electric fields. For pore formation giant unilamellar vesicles (GUVs) were used to avoid solvent and tension effects, and transport kinetics were measured by the entry of the impermeant fluorescent dye calcein. Because the timescale of electrical pulses needed to restructure bilayers into pores is much shorter than the time resolution of current techniques for membrane transport kinetics measurements, we measured the lifetimes of lipid bilayer electro-pores using systematic variation of the initial MD simulation conditions, whereas GUV transport kinetics were detected in response to a nanosecond-timescale variation in the applied electric pulse lifetimes and interpulse intervals.
Molecular transport after GUV permeabilization induced by multiple pulses is additive for interpulse intervals as short as 50 ns but not for 5–ns intervals, consistent with the 1050– ns lifetimes of electro-pores in MD simulations. Although the results were mostly consistent between GUV and MD simulations, the kinetics of ultrashort, electric field–induced permeabilization of GUVs were significantly different from published results in cells exposed to ultrashort (6 and 2 ns) electric fields, suggesting that cellular electro-poration involves additional structures and processes. The experimental data are consistent with the hypothesis that calcein-permeable lipid electro-pores in GUVs are created within a few nanoseconds and that most are annihilated within a few tens of nanoseconds, consistent with molecular simulations, but in contrast to the typical persistent electro-permeabilization (many seconds to minutes) observed in living cells. Furthermore, the magnitudes of the increases in intravesicular dye concentration are much smaller with nanosecond-pulsed electric fields than those observed in the presence of the pore-forming peptide melittin or exposure to influenza virus at a low pH. Nanosecond bipolar pulse cancellation, a phenomenon recently described in cells, was not observed in GUVs. The absence of persistent electro-permeabilization and nanosecond bipolar cancellation of GUVs suggests that the electro-permeabilization of cells involves structures and processes that go beyond transport through lipid pores and that models of electro-poration must be modified accordingly.
Determining the structure of the placental malaria vaccine candidate VAR2CSA
Women become more susceptible to malaria infection during pregnancy despite pre-existing immunity acquired from childhood, causing substantial risk of severe outcomes for the mother and her offspring. Placental malaria is caused by the accumulation of Plasmodium falciparum–infected erythrocytes in the placenta of pregnant women, resulting in high rates of maternal anemia, low birth weight, stillbirth, and spontaneous pregnancy loss. Each year, up to 200,000 infant deaths and 10,000 maternal deaths globally are attributed to malaria infection in pregnancy. The surge in deaths can be traced to the development of parasite resistance to the affordable drug chloroquine. Artemisinin, a newer, powerful, and affordable antimalarial drug, appeared promising at first, but resistant strains now abound. Vaccines have proven very difficult to produce, but focusing on women provides a strong basis for the development of vaccines to prevent placental malaria, because women naturally acquire resistance to placental malaria over successive pregnancies.
P. falciparum expresses a family of proteins, referred to as erythrocyte membrane protein 1 (PfEMP1), that are translocated to the surface of the infected erythrocyte to enable adherence to different host organs and to evade the host immune response. VAR2CSA (parasite-encoded variant surface antigens binding to chondroitin sulfate A), the leading placental malaria vaccine candidate, is a member of the PfEMP1 family that specifically binds to the syncytiotrophoblast surface receptor chondroitin sulfate A. The interaction facilitates placental sequestration of malaria parasites, leading to placental malaria. Given its large size (including a 310–kDa extracellular domain), production of VAR2CSA protein for vaccine development and scientific study has proved to be challenging. Furthermore, the highly polymorphic nature of the extracellular domain of VAR2CSA in parasite isolates may hinder the development of a strain-transcending vaccine. Lastly, vaccine-induced and naturally acquired immunity may differ in important ways that need to be carefully examined. CSA (chondroitin sulfate A) is presented by diverse cancer cells, and specific targeting of cells by VAR2CSA may become a viable approach for cancer treatment. We determined the Cryo-EM structures of the full-length ectodomain of VAR2CSA from P. falciparum strain NF54 in complex with CSA, and VAR2CSA from a second P. falciparum strain FCR3. VAR2CSA is composed of a stable core flanked by a flexible arm. CSA traverses the core domain by binding within two channels, and CSA binding does not induce major conformational changes in VAR2CSA. The CSA–binding elements are conserved across VAR2CSA variants and are flanked by polymorphic segments, suggesting immune selection outside the CSA–binding sites. The work thus provides paths for developing interventions against placental malaria and cancer.
Additional Funding
- Office of AIDS Research Innovative Research Proposal for Capital Equipment
Publications
- Chen CZ, Xu M, Pradhan M, Gorshkov K, Petersen JD, Straus MR, Zhu W, Shinn P, Guo H, Shen M, Klumpp-Thomas C, Michael SG, Zimmerberg J, Zheng W, Whittaker GR. Identifying SARS-CoV-2 entry inhibitors through drug repurposing screens of SARS-S and MERS-S pseudotyped particles. ACS Pharmacol Transl Sci 2020;3:1165–1175.
- Jain V, Shelby T, Patel T, Mekhedov E, Petersen JD, Zimmerberg J, Ranaweera A, Weliky DP, Dandawate P, Anant S, Sulthana S, Vasquez Y, Banerjee T, Santra S. A bimodal nanosensor for probing influenza fusion protein activity using magnetic relaxation. ACS Sens 2021;6:1899–1909.
- Sözer EB, Haldar S, Blank PS, Castellani F, Vernier PT, Zimmerberg J. Dye transport through bilayers agrees with lipid electropore molecular dynamics. Biophys J 2020;119:1724–1734.
- Garten M, Beck JR, Roth R, Tenkova-Heuser T, Heuser J, Istvan E, Bleck CKE, Goldberg DE, Zimmerberg J. Contacting domains segregate a lipid transporter from a solute transporter in the malarial host-parasite interface. Nat Commun 2020;11:3825.
- Ma R, Lian T, Huang R, Renn JP, Petersen JD, Zimmerberg J, Duffy PE, Tolia NH. Structural basis for placental malaria mediated by Plasmodium falciparum VAR2CSA. Nat Microbiol 2021;6:380–391.
Collaborators
- Sergei Akimov, PhD, Frumkin Institute of Physical Chemistry and Electrochemistry, Russian Academy of Sciences, Moscow, Russia
- Pasha Bashkirov, PhD, Frumkin Institute of Physical Chemistry and Electrochemistry, Russian Academy of Sciences, Moscow, Russia
- Oleg Batishchev, PhD, Frumkin Institute of Physical Chemistry and Electrochemistry, Russian Academy of Sciences, Moscow, Russia
- Christopher Karl Ernst Bleck, PhD, Electron Microscopy Core Facility, NHLBI, Bethesda, MD
- Catherine Z. Chen, PhD, National Center for Advancing Translational Sciences, Rockville, MD
- Patrick E. Duffy, PhD, Laboratory of Malaria Immunology and Vaccinology, NIAID, Rockville, MD
- Vadim Frolov, PhD, Universidad del País Vasco, Bilbao, Spain
- Timur Galimzyanov, PhD, Frumkin Institute of Physical Chemistry and Electrochemistry, Russian Academy of Sciences, Moscow, Russia
- Daniel Goldberg, MD, PhD, Washington University St. Louis, St. Louis, MO
- Samuel T. Hess, PhD, University of Maine, Orono, ME
- Ron W. Holz, MD, PhD, University of Michigan Medical School, Ann Arbor, MI
- Mary Kraft, PhD, University of Illinois at Urbana-Champaign, Urbana, IL
- Richard Pastor, PhD, Laboratory of Membrane Biophysics, NHLBI, Bethesda, MD
- Thomas S. Reese, MD, Laboratory of Neurobiology, NINDS, Bethesda, MD
- Santimukul Santra, PhD, Pittsburg State University, Pittsburg, KS
- Anna Shnyrova, PhD, Universidad del País Vasco, Bilbao, Spain
- Niraj H. Tolia, MD, PhD, Laboratory of Malaria Immunology and Vaccinology, NIAID, Bethesda, MD
- Peter K. Weber, PhD, Lawrence Livermore National Laboratory, Livermore, CA
- Gary R. Whittaker, PhD, College of Veterinary Medicine, Cornell University, Ithaca, NY
- Jack Yanovski, MD, PhD, Section on Growth and Obesity, NICHD, Bethesda, MD
- Wei Zheng, PhD, Division of Preclinical Innovation, NCATS, Bethesda, MD
Contact
For more information, email zimmerbj@mail.nih.gov or visit https://irp.nih.gov/pi/joshua-zimmerberg.