Phosphoinositide Messengers in Cellular Signaling and Trafficking

- Tamás Balla, MD, PhD, Head, Section on Molecular Signal Transduction
- Yeun Ju Kim, PhD, Staff Scientist
- Alejandro Alvarez-Prats, PhD, Postdoctoral Fellow
- Gerald Hammond, PhD, Postdoctoral Fellow
- Mira Sohn, PhD, Postdoctoral Fellow
- Nivedita Sengupta, PhD, Postdoctoral Fellow
- Ljubisa Vitkovic, PhD, Special Volunteer
We investigate signal transduction pathways that mediate the actions of hormones, growth factors, and neurotransmitters in mammalian cells, with special emphasis on the role of phosphoinositide-derived messengers. Phosphoinositides constitute a small fraction of the cellular phospholipids but play critical roles in the regulation of many signaling protein complexes that assemble on the surface of cellular membranes and are intracellular lipid messengers controlling a variety of cellular functions. Phosphoinositides regulate protein kinases and GTP–binding proteins as well as membrane transporters, including ion channels, thereby controlling many cellular processes such as proliferation, apoptosis, metabolism, cell migration, and differentiation. We focus on the phosphatidylinositol 4 (PtdIns4)–kinases (PI4Ks), a family of enzymes that catalyze the first committed step in polyphosphoinositide synthesis. Current work aims to: (1) understand the function and regulation of several PI4Ks involved in the control of cellular signaling and trafficking pathways; (2) find specific inhibitors for the individual PI4Ks; (3) define the molecular basis of PtdIns4P–regulated pathways through identification of PtdIns4P–interacting molecules; (4) develop tools to analyze inositol lipid dynamics in live cells; and (5) determine the importance of the lipid-protein interactions in the activation of cellular responses by G protein–coupled receptors and receptor tyrosine kinases.
EFR3 proteins are critical for G protein–coupled receptor re-sensitization.
We showed previously that significant amounts of phosphatidylinositol 4-phosphate (PI4P) can be found in the plasma membrane (PM) and that these pools are generated by the PI4KA enzyme. We set out to determine the mechanism by which PI4KA acts at the PM. Endogenous PI4K is detected in the ER, and GFP–tagged PI4KA is found in the cytosol but cannot be detected in the PM, even with TIRF microscopy. Yeast studies from Scott Emr's lab at Cornell University showed that two yeast proteins, Efr3p and Ypp1p, are required for the yeast ortholog of PI4KA, Stt4p, to localize to the PM. We thus investigated the role of the homologs of these proteins (EFR3A and EFR3B, and TTC7A and TTC7B) in localizing PI4KA to the PM in mammalian cells. We found that EFR3 proteins are palmitoylated at their N-termini and are localized to the PM and that their RNAi–mediated knock-down interferes with signaling of the G protein–coupled angiotensin II (AngII) receptors that activate phospholipase C (PLC) enzymes. While our studies reached their conclusions, Pietro De Camilli's group at the Yale School of Medicine reported on the role of the TTC7 and EFR3 proteins in localizing PI4KA in the PM. Our results were in agreement with those reported in their publication but also revealed an additional role of the EFR3 proteins, namely their critical role in the resensitization of the AngII receptors. Without the EFR3 proteins, AngII receptors rapidly uncoupled from the Gq protein and became desensitized. Our results explained the remarkable phenotype of a Drosophila mutant (rolling blackout, or rbo), which carries a mutated Efr3 gene and shows rapid desensitization of light-induced electrical responses in the eye. Whether the effects of Erf3 knock-down on GPCR resensitization are related to EFR3's role in supporting PI4KA function in the PM is a question that we are currently investigating.
Identification of Nir2 as a phosphatidylinostol and phosphatidic acid lipid transfer protein critical for maintaining signaling competence from phospholipase C–activating receptors
Another important achievement of this year’s research was the completion of a study that addressed the question of how structurally important lipids are exchanged between the endoplasmic reticulum (ER) and the plasma membrane (PM) during the actions of hormones and neurotransmitters. Our studies focused on the question of how phosphatidylinositol (PI), the precursor lipid of the regulatory phosphoinositides, is transported from the ER to other membranes. We tested all known PI transfer proteins (PITPs) by RNA–mediated knock-down and found that only Nir2 had a profound effect on phosphoinositide levels. Nir2 is a large PITP, first described in Drosophila as the RdgB mutant, which develops light-induced retinal degeneration. We found that Nir2–depleted cells' ability to supply the PM with PI was compromised, as was their PI synthesis, owing to a defect in phosphatidic acid (PA) transport from the PM to the ER. We also found that Nir2 was enriched in ER–PM contact sites after stimulation of phospholipase C (PLC) enzymes and that the Nir2 protein was anchored to the ER via interaction with VAP-A and VAP-B proteins. The studies answered a long-standing question of how the lipid products of PLC activation are recycled to maintain the signaling competence of cells under chronic stimulation. Given that some familial forms of amyotrophic lateral sclerosis (ALS or Lou-Gehrig’s disease) are caused by mutations within the VAP-B protein, we also tested these mutants for their ability to interact with the Nir2 protein. We found that some disease-causing mutations largely interrupted the interaction between Nir2 and VAB-B, while others had more moderate effects. The studies may give us further clues regarding the molecular details of the processes that are critical for the pathology of ALS.
Click image to enlarge.
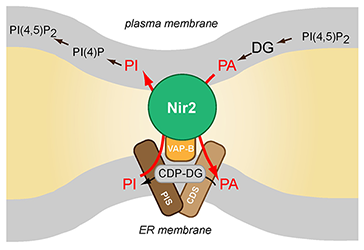
Schematics of Nir2 function in ER-PM contact sites
The Nir2 protein is a lipid transporter that moves phosphatidylinositol (PI) from the endoplasmic reticulum (ER) to the plasma membrane (PM) while transferring phosphatidic acid (PA) in the other direction. Nir2 translocates to the ER-PM contact sites when cells are stimulated via receptors that activate phospholipase C (PLC) enzymes to fulfill the protein's lipid exchange function. Therefore, Nir2 helps maintain plasma membrane lipid identity and signaling competence when phosphoinositides are rapidly consumed during receptor-mediated activation of PLC enzymes.
Analysis of the fatty acid preference of cytidine-diphosphate-diacylglycerol synthase (CDS) enzymes
Cytidine-diphosphate(CDP)-diacylglycerol(DG) synthase (CDS) enzymes catalyze the formation of CDP-DG from phosphatidic acid (PA) and cytosine triphosphate (CTP) and are found in a wide range of organisms, from prokaryotes to humans. The reaction is the first enzymatic step in generating phosphatidylinositol (PI), which is the precursor of phosphoinositides. Cells have multiple forms of CDS enzymes; CDS1 and CDS2 are multi-pass transmembrane proteins located in the ER, while the mitochondrial CDS, called Tam41 is a soluble protein associated with the inner mitochondrial membrane. In this set of studies, performed in collaboration with Richard Epand's group, we compared the human CDS1 and CDS2 enzymes' selective preference for PA with special fatty acid side-chain composition. The importance of this question lies in the fact that the fatty acid side chain composition of newly synthesized PA is dipalmitoyl, whereas those generated from hydrolysis of phosphoinositides following phospholipase C activation are 1-stearoyl, 2-arachidonyl. Therefore, the analysis was designed to investigate whether the two CDS enzymes show any preference for the PA that originates from receptor-mediated breakdown of phosphoinositides. The studies revealed that the CDS2 enzyme prefers the 1-stearoyl, 2-arachidonyl version of PA, while CDS1 can use any form of PA without distinction. Although both enzymes are located in the ER, the functional difference suggests a metabolic channeling of PA toward the various phospholipid metabolic pathways by the two CDS enzymes.
Publications
- Hammond GR, Balla T. Polyphosphoinositide binding domains: key to inositol lipid biology. Biochim Biophys Acta 2015; 1851(6):746-758.
- Bojjireddy N, Guzman-Hernandez ML, Reinhard NR, Jovic M, Balla T. EFR3s are palmitoylated plasma membrane proteins that control responsiveness to G-protein-coupled receptors. J Cell Sci 2015; 128:118-128.
- D'Souza K, Kim YJ, Balla T, Epand RM. Distinct properties of the two isoforms of CDP-diacylglycerol synthase. Biochemistry 2014; 53:7358-7367.
- Jeschke A, Zehethofer N, Lindner B, Krupp J, Schwudke D, Haneburger I, Jovic M, Backer JM, Balla T, Hilbi H, Haas A. Phosphatidylinositol 4-phosphate and phosphatidylinositol 3-phosphate regulate phagolysosome biogenesis. Proc Natl Acad Sci USA 2015; 112:4636-4641.
- Kim YJ, Guzman-Hernandez ML, Wisniewski E, Balla T. Phosphatidylinositol-phosphatidic acid exchange by Nir2 at ER-PM contact sites maintains phosphoinositide signaling competence. Dev Cell 2015; 33:549-561.
Collaborators
- Evžen Boura, PhD, Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic, Prague, Czech Republic
- Richard Epand, PhD, McMaster University, Hamilton, Ontario, Canada
- Albert Haas, PhD, Institut für Zellbiologie, Universität Bonn, Bonn, Germany
- Péter Várnai, MD, PhD, Semmelweis University, Faculty of Medicine, Budapest, Hungary
Contact
For more information, email ballat@mail.nih.gov or visit https://www.nichd.nih.gov/about/staff/Pages/bio.aspx?nih_id=0010183740.


