Deciphering the Virulence Program of Legionella pneumophila

- Matthias Machner, PhD, Head, Unit on Microbial Pathogenesis
- Eric Cheng, PhD, Postdoctoral Fellow
- Nicole Ellis, PhD, Postdoctoral Fellow
- Byoungkwan Kim, PhD, Postdoctoral Fellow
- Pei-Chung Lee, PhD, Postdoctoral Fellow
- Yi-han Lin, PhD, Postdoctoral Fellow
- Alexandra Doms, BA, Postbaccalaureate Student
- D'anna Nelson, BA, Postbaccalaureate Student
Our main research goal is to obtain mechanistic insight into the virulence strategies of microbial pathogens. As a model organism we use the bacterium Legionella pneumophila, the causative agent of a potentially fatal respiratory infection known as Legionnaires' disease. Contrary to what its name may imply, Legionnaires’ disease occurs in individuals of all ages, including children who receive respiratory therapy, newborns who recently underwent surgery or under-water birth, and children who are immune-compromised. We are committed to the in-depth analysis of mechanisms that allow L. pneumophila to exploit the human host and cause disease. Insights gained from these studies will ultimately improve our ability to better diagnose, prevent, and fight Legionnaires’ disease and related illnesses, thereby contributing to the success of NICHD’s mission.
Upon inhalation of contaminated water droplets, L. pneumophila enters the lung and is phagocytosed (taken up) by alveolar macrophages, specialized immune cells. Instead of being degraded by these cells, the pathogen establishes a protective membrane compartment, the Legionella-containing vacuole (LCV). Within this intravacuolar niche, L. pneumophila can replicate to high numbers before it kills the host cell and infects neighboring cells.
Intracellular survival of L. pneumophila depends on the activity of more than 300 proteins, or effectors, that are injected into the host cell, where they create conditions favorable for infection. L. pneumophila mutants that are defective in effector protein delivery fail to escape endolysosomal degradation, underscoring the key role of microbial effectors for bacterial virulence. Our goal is to obtain a detailed mechanistic insight into the regulation and function of L. pneumophila effectors by investigating host-pathogen interactions at a molecular, cellular, and structural level. Deciphering the virulence program of this dangerous pathogen will set the stage for the development of novel therapeutics aimed at treating or preventing Legionnaires' disease and related illnesses.
L. pneumophila exploits host-mediated S-palmitoylation for effector localization.
Although the existence within a camouflaged membrane-enclosed compartment provides several benefits to L. pneumophila, it also creates certain challenges. For example, how are microbial effector proteins, once they have been delivered into the host cytosol, directed towards the correct host target or organelle where they can then exhibit their function? Thus far, only few targeting mechanisms have been described, including the binding of effectors either to specific proteins or to phospholipids (References 1 and 2).
Our study of the effector protein GobX from L. pneumophila revealed yet another targeting strategy that exploits a form of lipidation called S-palmitoylation (Figure 1A) (Reference 3). S-palmitoylation is the covalent attachment of palmitic acid, a hydrophobic C-16 carbohydrate chain, to cysteine residues of proteins. The reaction is catalyzed by eukaryotic palmitoyl transferases (PATs), also known as DHHC proteins owing to a conserved catalytic aspartate-histidine-histidine-cysteine motif. More than 500 mammalian proteins have been shown to be covalently modified with palmitic acid, making S-palmitoylation a post-translational modification of great importance. Despite the abundance of S-palmitoylated proteins in eukaryotic cells, the consensus sequence for this post-translational modification had remained unclear.
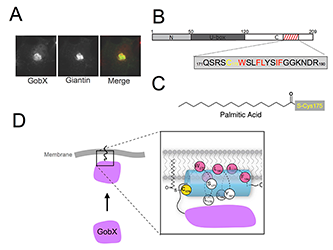
We discovered that the L. pneumophila GobX specifically localizes to the Golgi compartment of host cells in a process that requires host-mediated S-palmitoylation. A surprisingly short stretch of 20 amino acids (aa) within the C-terminal domain of GobX was sufficient for Golgi targeting (Figure 1B). Upon closer examination, we identified a cysteine residue at position 175 (C175) that was essential for Golgi localization. Using metabolic labeling and Click chemistry, we discovered that C175 was the target of host cell–mediated S-palmitoylation (Figure 1C). Substitution of C175 by either serine or alanine prevented S-palmitoylation and, thus, proper intracellular targeting of GobX. Notably, C175 was not the only residue within the 20 aa region of GobX that was essential for Golgi localization. Using site-directed mutagenesis, we identified several additional residues that were equally important for S-palmitoylation (highlighted red in Figure 1B). Interestingly, each of these important residues was hydrophobic in nature and positioned at one side of a predicted alpha helix in GobX, giving it an amphipathic character. We propose that the amphipathic helix can peripherally associate with Golgi membranes, thus bringing C175 into proximity with the DHHC active site of Golgi-resident PATs in order for S-palmitoylation to occur (Figure 1D).
Our in-depth analysis of GobX not only revealed a novel targeting mechanism for L. pneumophila effectors, but also provided the first evidence that the consensus motif for S-palmitoylation may be structure- rather than sequence-based (the amphipathic nature of the helix is more important than its exact sequence), explaining why no consensus sequence for S-palmitoylation had previously been identified in eukaryotes.
Click image to enlarge.
Figure 1. GobX exploits S-palmitoylation for Golgi localization.
A. Intracellular localization of GobX. Transiently transfected COS-1 cells producing GFP-GobX were labeled with antibody directed against the Golgi marker giantin. The merged image (right) shows GobX in green and giantin in red. B. Schematic illustration of the domain organization of GobX. Numbers indicate amino acid positions. The minimal Golgi-localizing fragment (residue 171–190) is indicated by the hatched box. The amino acid sequence of this region is shown in single letter code, with C175 highlighted in yellow and hydrophobic residues of the amphipathic helix in red. C. Chemical structure of palmitic acid covalently attached to C175. D. Model of membrane association and S-palmitoylation of GobX by PATs. Color coding as in (B).
Host-pathogen interaction profiling using self-assembling human protein arrays
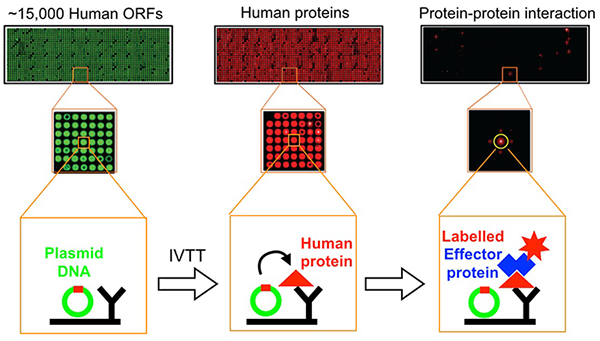
Despite many years of intense studies by several groups, L. pneumophila effectors of unknown function vastly outnumber those that have been well characterized. The identification of host targets has remained particularly challenging owing to the lack of simple detection tools that avoid abundance biases while providing an open format for experimental modifications. Together with the group of Joshua LaBaer, we established an improved protein-protein interaction platform called Nucleic Acid-Programmable Protein Array (NAPPA) (Reference 4). For this array, thousands of genes encoding tagged human bait proteins are printed on an aminosilane-coated slide (Figure 2). At the time of assay, the proteins are freshly synthesized through in vitro transcription/translation (IVTT) and displayed in situ using co-spotted anti-tag antibodies.
We developed an improved NAPPA by introducing the HaloTag (Promega) at the C-terminus of the bacterial query protein. HaloTag is a modified haloalkane dehalogenase designed to covalently bind to synthetic Halo-ligands (haloalkanes). Once applied to NAPPA, binding of a HaloTag query protein to its interactor(s) can be specifically detected among thousands of proteins using an Alexa660-labeled Halo-ligand. In a proof-of-concept study, we probed the NAPPA with the L. pneumophila effectors SidM or LidA and identified most of the known targets but also potential novel interaction candidates, a subset of which we confirmed in independent in vitro pull-down and in vivo cell-based assays, thus providing further insight into how these effectors may discriminate between different host Rab GTPases.
In addition, by combining NAPPA with a chemical labeling technique called Click (Copper(I)-catalyzed azide-alkyne Huisgen cycloaddition) chemistry (in collaboration with Howard Hang), we generated a modified screening platform capable of identifying human targets of post-translational modifications such as AMPylation (adenylylation) (Reference 4). We identified the previously reported AMPylation targets of SidM, namely Rab1 and Rab35, and also discovered several novel AMPylation targets, whose contribution to a successful L. pneumophila infection has yet to be determined. Our improved NAPPA platform shows broad applicability and can be adapted for the high-throughput analysis of effectors from a variety of pathogens.
Click image to enlarge.
Figure 2. Flow scheme of NAPPA fabrication and protein interaction assay
NAPPA arrays are converted from DNA to protein through in vitro transcription/translation. Newly synthesized tagged human proteins are captured on the array by a tag-specific antibody, and binding of HaloTag-query protein to its target on NAPPA is detected using Alexa660-labeled Halo-ligand. Plasmid cDNA on the array is stained green, proteins red.
Additional Funding
- NICHD Director's Challenge Innovation Award
Publications
- Gaspar AH, Machner MP. VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion. Proc Natl Acad Sci USA 2014; 111:4560-4565.
- Lucas M, Gaspar AH, Pallara C, Rojas AL, Fernández-Recio J, Machner MP, Hierro A. Structural basis for the recruitment and activation of the Legionella phospholipase VipD by the host GTPase Rab5. Proc Natl Acad Sci USA 2014; 111:E3514-3523.
- Lin YH, Doms AG, Cheng E, Kim B, Evans TR, Machner MP. Host cell-catalyzed S-palmitoylation mediates Golgi targeting of the Legionella ubiquitin ligase GobX. J Biol Chem 2015; 290:25766-25781.
- Yu X, Decker KB, Baker K, Neunuebel MR, Graves M, Westcott N, Hang H, LaBaer J, Qiu J, Machner MP. Host-pathogen interaction profiling using self-assembling human protein arrays. J Proteome Res 2015; 14:1920-1936.
Collaborators
- Howard Hang, PhD, The Rockefeller University, New York, NY
- Aitor Hierro, PhD, CIC bioGUNE Institute, Bilbao, Spain
- Joshua LaBaer, MD, PhD, Virginia G. Piper Center for Personalized Diagnostics, Arizona State University, Tempe, AZ
Contact
For more information, email machnerm@mail.nih.gov or visit http://machnerlab.nichd.nih.gov.


