Chromatin Remodeling and Gene Activation

- David J. Clark, PhD, Head, Section on Chromatin and Gene Expression
- Peter Eriksson, PhD, Staff Scientist
- Razvan V. Chereji, PhD, Visiting Fellow
- Josefina Ocampo, PhD, Visiting Fellow
- Sean C. Clark, MS, Postbaccalaureate Fellow
- Allison F. Dennis, BS, Graduate Student
Aberrant gene regulation is the basis of many disease states. Our main objective is to understand how genes are activated for transcription in the context of chromatin structure. Chromatin is not just a packaging system for DNA in eukaryotic cells but also participates in gene regulation. The structural subunit of chromatin is the nucleosome, which contains nearly two turns of DNA coiled around a central core histone octamer. Nucleosomes are generally quite regularly spaced along the DNA, like beads on a string. Gene regulation involves either attenuation of the inherently repressive properties of nucleosomes to facilitate gene expression or enhancement of those properties to ensure complete repression. These events are choreographed by DNA sequence–specific transcription factors (activators and repressors) and chromatin-remodeling complexes. The latter can be divided into two groups: histone- or DNA–modifying enzymes that implement the 'epigenetic code', and ATP–dependent remodeling machines that move or displace nucleosomes. We are exploiting the new high-throughput technologies to obtain genome-wide maps of nucleosomes, chromatin-remodeling complexes and RNA polymerase II in budding yeast to determine what happens to nucleosomes on genes when they are activated. We find that transcription results in disruption and loss of some nucleosomes on the gene with re-positioning of the remaining nucleosomes. The current objectives of our yeast studies are to: (1) determine the roles of various chromatin-remodeling complexes (RSC, SWI/SNF, ISW1, ISW2, and CHD1) in chromatin organization and gene expression. They are important because genes encoding subunits of some of these enzymes are often mutated in various cancers and in some developmental diseases; (2) understand the dynamics of transcription by RNA polymerase II through chromatin in vivo, with an emphasis on potential regulation of the dissociation of polymerase after transcription.
In addition, through collaborations, we are now extending our studies of chromatin remodeling from yeast to mouse. Mammalian systems are much more complex and challenging than yeast. We are investigating the effects of dexamethasone on mouse cell chromatin in a collaboration with the Hager lab. We are also comparing the chromatin structures of neurons, oligodendrocytes, and astrocytes in a collaboration with the Fields lab.
Disruption of nucleosomes during heavy transcription
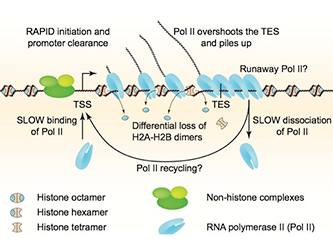
The histone octamer contains an H3-H4 tetramer and two H2A-H2B dimers. Gene activation is associated with chromatin disruption: a wider nucleosome-depleted region (NDR) at the promoter and reduced nucleosome occupancy over the coding region. We examined the nature of disrupted chromatin after induction, using MNase-seq to map nucleosomes and sub-nucleosomes and a refined high-resolution ChIP-seq method to map histones H4 and H2B and RNA polymerase II (Pol II) genome-wide. Over coding regions, induced genes show a differential loss of H2B relative to H4, which correlates with Pol II density and the appearance of sub-nucleosomes. After induction, Pol II is surprisingly low at the promoter, but accumulates on the gene and downstream of the termination site, implying that dissociation is slow. Thus, induction-dependent chromatin disruption reflects both eviction of H2A-H2B dimers from nucleosomes and the presence of queued Pol II elongation complexes (Figure 1). We propose that slow Pol II dissociation after transcription is a major factor in chromatin disruption and that it may be of critical importance in gene regulation (Reference 1), a hypothesis that we are currently testing.
Click image to enlarge.
Figure 1. Chromatin structure of a heavily transcribed gene
Model: RNA polymerase II (Pol II) binds near the transcription start site (TSS) and clears the promoter rapidly. Pol II levels over the remainder of the gene are high and correlate with some nucleosome displacement and some loss of histone H2A-H2B dimers, resulting in sub-nucleosomal particles (tetrasomes and hexasomes). Pol II overshoots the transcript end site (TES) and accumulates downstream owing to slow dissociation from DNA, resulting in a queue of polymerases behind it. Slow dissociation might facilitate transfer of Pol II back to the promoter (‘recycling’), thereby increasing the transcription rate.
Novel nucleosomal particles containing core histones and linker DNA but no histone H1
Eukaryotic chromosomal DNA is assembled into regularly spaced nucleosomes, which play a central role in gene regulation by determining accessibility of control regions. The nucleosome contains about 147 bp of DNA wrapped approximately 1.7 times around the core histone octamer. The linker histone H1 binds both to the nucleosome, sealing the DNA coils, and to the linker DNA between nucleosomes, directing chromatin folding. Micrococcal nuclease (MNase) digests the linker to yield the chromatosome, containing H1 and about 160 bp, and then converts it to a core particle, containing about 147 bp and no H1. Sequencing of nucleosomal DNA obtained after MNase digestion (MNase-seq) generates genome-wide nucleosome maps that are important for understanding gene regulation. We developed an improved MNase-seq method involving simultaneous digestion with exonuclease III, which removes linker DNA. Remarkably, we discovered two novel intermediate particles containing 154 or 161 bp, corresponding to 7 bp protruding from one or both sides of the nucleosome core (Reference 2). The particles are detected in yeast lacking H1 and in H1–depleted mouse chromatin. They can be reconstituted in vitro using purified core histones and DNA. We propose that these "proto-chromatosomes" are fundamental chromatin subunits, which include the H1 binding site and influence nucleosome spacing independently of H1 (Figure 2).
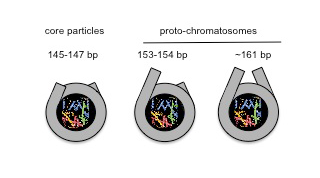
Figure 2. Proto-chromatosomes form the scaffold for linker histone binding.
Views of the nucleosome core particle (147 bp) and proto-chromatosomes from above, drawn approximately to scale, based on the nucleosome crystal structure. The final 10 bp on each side of the nucleosome core are almost straight, projecting a short distance out of the particle. Proto-chromatosomes are shown with an extra 7 bp on one side (154 bp) or both sides (161 bp) with a continuing straight trajectory. Linker histone (H1) binds to the 161-bp proto-chromatosome, interacting with the protruding DNA on both sides and with the central turn of DNA within the nucleosome.
The ISW1 and CHD1 ATP–dependent chromatin remodelers compete to set nucleosome spacing in vivo.
ATP–dependent chromatin remodeling machines play a central role in gene regulation by manipulating chromatin structure. Most genes have a nucleosome-depleted region at the promoter and an array of regularly spaced nucleosomes phased relative to the transcription start site. The three nucleosome spacing enzymes in yeast (CHD1, ISW1, and ISW2) catalyze the formation of arrays with different average spacing in vitro (around 160, 175, and 200 bp, respectively). We used MNase-seq to show that nucleosome spacing in vivo reflects competition between CHD1 and ISW1 to set the spacing on individual genes, such that CHD1 dominates genes with shorter spacing and ISW1 dominates genes with longer spacing. ISW2 plays a minor role limited to inactive genes. Active genes exhibit extreme spacing, both very short and very long. H1 binding increases with spacing, suggesting that arrays with shorter spacing, created by CHD1, may exclude H1 because the linker is too short for tight binding (Ocampo J, Chereji RV, Eriksson P, Clark DJ, manuscript submitted).
Genome-wide cooperation by HAT Gcn5, remodeler SWI/SNF, and chaperone Ydj1 in promoter nucleosome eviction and transcriptional activation
As mentioned above, gene activation generally involves nucleosome removal and/or nucleosome shifts in the promoter region. The process of nucleosome assembly and disassembly at promoters involves histone chaperones, chromatin-remodeling complexes, and histone acetyltransferases. We investigated whether these co-factors function ubiquitously and what effect nucleosome eviction has on transcription genome-wide. We performed chromatin immunoprecipitation (ChIP-seq) experiments to detect histone H3 and Pol II in mutants lacking single or multiple co-factors (the SWI/SNF remodeling complex, the Gcn5 histone acetyltransferase, and the Ydj1 histone chaperone). We focused on about 200 genes regulated by the transcription activator Gcn4. After induction, 70 of these genes show significantly reduced H3 occupancy at their promoters. All three co-factors examined play a role in H3 eviction at most, but not all, Gcn4 target promoters, with Gcn5 playing the greatest role and Ydj1 the least. In fact, the three co-factors cooperate to various extents in H3 eviction at virtually all yeast promoters. Reduced H3 eviction in co-factor mutants is correlated with reduced Pol II occupancy on Gcn4–regulated genes and on the most highly expressed constitutive genes, but the relative Pol II levels at most genes are unaffected. Our observations indicate that promoter nucleosome eviction is important for heavy transcription of highly expressed genes, but that other steps in gene activation are more important for most other yeast genes (Qiu H, Hu C, Chereji R, Cole HA, Rawal Y, Clark DJ, Hinnebusch AG, manuscript submitted).
Chromatin remodeling in neurons and oligodendroglia
The aim of this project, which is being conduction in collaboration with Douglas Fields and Philip Lee, is to determine the relationship between chromatin organization and gene expression in dorsal root ganglion neurons and oligodendroglial cells. Neuronal chromatin is remarkably atypical: the linker DNA between nucleosomes is much shorter than in other cell types, including oligodendroglial cells, and neurons are deficient in histone H1. The global organization of neuronal chromatin is therefore very different from other cells, but the significance of this organization is unknown. We are testing the relationship between global chromatin organization and transcription. We are mapping nucleosomes genome-wide in isolated mouse neurons and in oligodendroglial cells, using paired-end sequencing, and are measuring gene expression by RNAseq. We will correlate their chromatin structures with transcriptional activity. The next step is to address the roles of ATP–dependent chromatin remodelers in neuronal and oligodendroglial chromatin structure. The experiments should provide insight into why neuronal chromatin organization is so different from other cells, including glia.
Publications
- Cole HA, Ocampo J, Iben JR, Chereji RV, Clark DJ. Heavy transcription of yeast genes correlates with differential loss of histone H2B relative to H4 and queued RNA polymerases. Nucleic Acids Res 2014; 42:12512-12522.
- Cole HA, Cui F, Ocampo J, Burke TL, Nikitina T, Nagarajavel V, Kotomura N, Zhurkin V, Clark DJ. Novel nucleosomal particles containing core histones and linker DNA but no histone H1. Nucleic Acids Res 2015; 44(2):573-81.
Collaborators
- Feng Cui, PhD, Rochester Institute of Technology, Rochester, NY
- Douglas Fields, PhD, Section on Nervous System Development and Plasticity, NICHD, Bethesda, MD
- Gordon L. Hager, PhD, Laboratory of Receptor Biology and Gene Expression, Center for Cancer Research, NCI, Bethesda, MD
- Alan G. Hinnebusch, PhD, Program in Cellular Regulation and Metabolism, NICHD, Bethesda, MD
- James Iben, PhD, Program in Genomics of Differentiation, NICHD, Bethesda, MD
- Philip Lee, PhD, Section on Nervous System Development and Plasticity, NICHD, Bethesda, MD
- Vasily M. Studitsky, PhD, Fox Chase Cancer Center, Temple University Health System, Philadelphia, PA
- Victor Zhurkin, PhD, Laboratory of Cell Biology, Center for Cancer Research, NCI, Bethesda, MD
Contact
For more information, email clarkda@mail.nih.gov or visit http://clarklab.nichd.nih.gov.


