Pineal Gland, Chronobiology, Neurotranscriptomics and Neuroepigenetics

- David C. Klein, PhD, Head, Section on Neuroendocrinology
- Cong Fu, PhD, Guest Researcher
Among the most important advances made by the Section are discoveries that resulted in the recognition of the suprachiasmatic nucleus as the site of the master oscillator controlling circadian biology in vertebrates; and the identifcation of the enzyme that serves to control melatonin production, the 'timezyme' arylalkylamine N-acetyltransferase (AANAT). The work has broad implications for vertebrate biology and is of special interest to clinical scientists studying human diseases related to circadian rhythms, including endocrine pathologies, sleep and mood disorders, and deficiencies in alertness. Other areas studied include the characterization of the transcriptome of the vertebrate pineal gland and the regulation of daily and developmental changes in the transcriptome of the pineal gland and of other tissues.
The 'timezyme' arylalkylamine N-acetyltransferase
The penultimate enzyme in the melatonin synthesis pathway is AANAT (Figure 1). We found that the enzyme is critical to the control of the rhythm in melatonin synthesis. In all species examined to date, the large increase in melatonin synthesis at night is a highly conserved feature of vertebrate physiology.
Click image to enlarge.
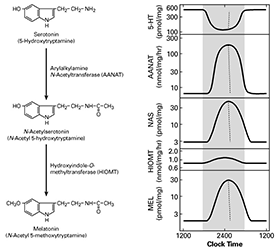
Figure 1. Daily rhythm in indole metabolism in the pineal gland
The daily rhythm in circulating melatonin production reflects the increased production in the pineal gland, as depicted here. During the day, melatonin production is low owing to low levels of AANAT. At night, AANAT activity rises, resulting in an increase in N-acetylserotonin. The increase in N-acetylserotonin raises melatonin production by a mass action effect; the level of the last enzyme in melatonin synthesis is constant. Changes in melatonin synthesis cause parallel changes in melatonin release. Circulating melatonin is rapidly destroyed in the liver, which allows changes in production and release to be immediately reflected in changes in circulating melatonin.
The transcriptional mechanisms that control expression of the Aanat gene in rodents and the chicken include interactions of cyclic AMP–response elements in the promoter region; other response elements in the promoter, including the E-box and photoreceptor conserved elements, appear to determine the marked tissue-specific pattern of expression of the gene, which is limited to the pineal gland and retina. We determined that, in rodents, expression of Aanat rises about 100-fold at night; however, this feature of regulation is not highly conserved. For example, it was discovered that there is little or no nocturnal increase in Aanat expression in ungulates or nonhuman primates, in contrast to rodents, birds, and fish. Thus, transcriptional control mechanisms are not important for the regulation of melatonin synthesis in all vertebrates.
Whereas transcriptional regulation is not highly conserved, post-translational control is and is mediated by cyclic AMP and involves phosphorylation of AANAT at C- and N-terminal phosphorylation sites by cyclic AMP–dependent protein kinase. This leads to formation of a reversible complex with 14-3-3 protein, in which the enzyme is stabilized and activated, as indicated by structural, in vivo, and in vitro studies. When cyclic AMP levels fall, AANAT is dephosphorylated and destroyed by the proteasome, causing enzyme activity and melatonin production to fall in parallel, with a halving time of about 3 to 4 minutes.
The form of AANAT found in the pineal gland evolved very rapidly approximately 500 million years ago from an enzyme dedicated to detoxification; this ancestor lacked the regulatory and catalytic features that are characteristic of the regulation and production of melatonin in the pineal gland. The evolutionary change—termed neo-functionalization—coincided with the appearance of the pineal gland and lateral eyes. The findings support the hypothesis that lateral eyes and the pineal gland evolved from a common primitive photodetector, which was facilitated by a primitive form of AANAT, which served to enhance photodetection by removing toxic arylalkylamines. The elevated activity at night is likely to have enhanced the ability to detect low levels of light. Subsequently, one of the downstream products of acetylation, melatonin, became a unique and valuable signal of night time—a time-keeping signal—which promoted the independent evolution of eyes, dedicated to image detection, and the pineal gland, dedicated to melatonin production and time keeping.
Neuroepigenetics: global control of daily changes in pineal gene expression are regulated by an adrenergic/cyclic AMP mechanism.
Transcriptome profiling
Several projects aimed to obtain a global picture of differences in gene expression that occur on a night/day basis and to identify genes that are highly enriched in the pineal gland. In collaborative work, we characterized pineal gene expression in the zebrafish, rat, mouse, chicken, and rhesus monkey as a function of time of day and of development. We used transcriptome profiling to analyze the pineal gland, by employing the most advanced profiling methods, which now include RNA sequencing, an effort that resulted in the identification of a set of genes that are highly expressed in the pineal gland. Some of the genes are expressed at high levels during the day and night; in other cases there are large changes in expression. The changes are seen in thousands of genes; of special interest are those with 10 to 100-fold changes in transcript abundance. Studies in the rat indicate that these are under neural control (Figures 2 and 3). This line of investigation has led to a rapid increase in knowledge of the biochemical profile, conserved across species, of the pineal gland. It is also pointing to new transcriptional pathways controlled by previously unrecognized transcription factors. Analysis of the transcription factors and the promoters of genes that are upregulated at night or highly expressed in the pineal gland will lead to the construction of a regulatory network that describes the cascade of transcription factors underlying the control of pineal gene expression. Our results permit a full-scale biochemical and physiological analysis of the control of genes in the pineal gland. The RNA sequencing work on the pineal transcriptome that clearly revealed a broad effect of neural/cyclic AMP regulation of the pineal gland (Figure 3) led to the creation of a new title for this line of investigation: neurotranscriptomics.
Click image to enlarge.
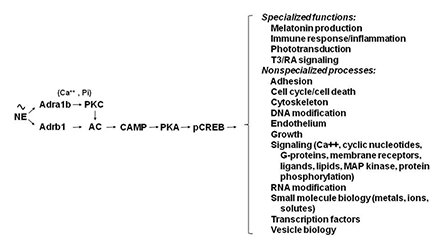
Figure 2. Neural regulation of gene expression in the rat pineal gland
Sympathetic nerves release norepinephrine (NE), which binds to beta1-adrenergic receptors (Adrb1) and to alpha1b-adrenergic receptors (Adra1b). NE acts through the Adrb1 receptor to partially activate adenylate cyclase (AC) and through Adra1b to potentiate activation of the Adrb1 receptor. The interaction involves Adra1b–dependent activation of Ca2+ and phosphatidylinositol (Pi) activation of protein kinase C (PKC), which enhances Adrb1 stimulation of AC. The resulting increase in cyclic AMP (CAMP) leads to activation of protein kinase A (PKA), which phosphorylates cyclic AMP response element–binding protein (CREB). The consequence of this is a global shift in gene expression. The transcription factor profile unique to this tissue establishes which genes are regulated by CREB phosphorylation.
Click image to enlarge.
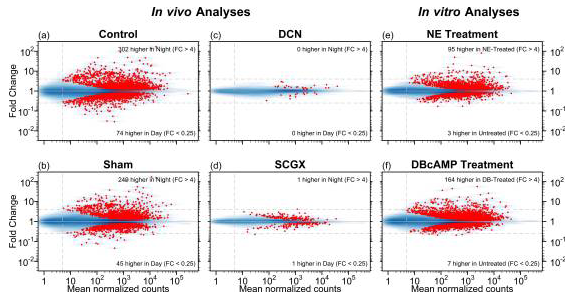
Figure 3. MA plots for six differential expression analyses
The MA plots [visual representation of two-channel DNA microarray gene expression data that has been transformed onto the M (log ratios) and A (mean average) scale] display the mean normalized read-pair counts (x-axis) versus the estimated fold change (y-axis), on a log-log scale for four in vivo analyses and two in vitro analyses. The blue shading indicates the density of genes, and each red point represents a gene with statistically significant differential expression. Dashed horizontal lines mark four-fold changes in both directions, dashed vertical line indicates minimum abundance threshold for the statistical tests. The four in vivo analyses compared night and day time points in adult rats for the following groups: (a) no surgery (Control); (b) neonatal sham surgery (Sham); (c) neonatal superior cervical ganglia decentralization (DCN); (d) neonatal superior cervical ganglionectomy (SCGX). The two in vitro analyses compared treated/untreated pineal glands: (e) norepinephrine-treated (NE) vs. untreated; and (f) dibutyryl-cyclic-AMP–treated (DBcAMP) vs. untreated (Reference 1).
Long noncoding RNAs
RNA sequencing identified a set of highly tissue-specific and rhythmically expressed non-coding RNAs. The transcripts range in size from approximately 200 bp to over 10,000 bp and vary in their genomic location from genetic deserts to being transcribed from introns of known genes. In at least one case, a long noncoding RNA overlaps a known gene and, in another, the long noncoding RNA is transcribed from the strand of DNA that is opposite to that encoding a known gene. The expression of these interesting molecules is regulated by the same neural system that controls coding transcripts. The transcripts may function in biological regulation through interaction with DNA or proteins.
MicroRNAs (miRNAs)
miRNAs play a broad range of roles in biological regulation. We profiled rat pineal miRNAs for the first time and evaluated their importance by focusing on melatonin synthesis, the main function of the pineal gland. Massively parallel sequencing and related methods revealed that the miRNA population is dominated by a small group of miRNAs as follows: about 75% is accounted for by 15 miRNAs; miR-182 represents 28%. In addition to miR-182, miR-183 and miR-96 are also highly enriched in the pineal gland, a distinctive pattern that is also found in the retina. The work also identified previously unrecognized miRNAs and other small noncoding RNAs. Pineal miRNAs do not exhibit a marked night/day differences in abundance with few exceptions (e.g., two-fold night/day differences in the abundance of miR-96 and miR-182), in sharp contrast to the dynamic 24-hour pattern that characterizes the pineal transcriptome. During development, the abundance of miRNAs most enriched in the pineal gland increases; however, there is a marked decline in at least one, miR-483. Based on the following observations, miR-483 is a likely regulator of melatonin synthesis: it inhibits the melatonin synthesis of pinealocytes in culture; it acts via predicted binding sites in the 3′ UTR of Aanat mRNA; and it exhibits the reverse developmental profile to that of Aanat transcripts. Additionally, a miR-483–targeted antagonist increased melatonin synthesis in neonatal pinealocytes. The observations support the hypothesis that miR-483 suppresses Aanat mRNA levels during development and that the developmental decline in miR-483 abundance promotes melatonin synthesis.
New element in biological timing
Studies of the zebrafish pineal gland identified a new element in chronobiology, the rhythmic gene camk1gb (encoding calcium/calmodulin-dependent protein kinase 1Gb), which is driven by the biological clock and links the clock with locomotor activity. This is important to an understanding of circadian control mechanisms in vertebrates.
Impact of RNA sequencing
The Section has been successful in promoting the use of RNA sequencing in programs not involved in pineal research, including work with other investigators at the NIH and elsewhere, such as studies on pain pathways, the hypothalamus, and the pituitary gland. In the latter case, RNA sequencing and our subsequent analysis revealed that the hypothalamic peptide GnRH induces a greater than 600-fold increase in expression of dentin matrix protein-1 (Kucka M et al., Mol Endocrinol 2013;27:1840-1850). The finding provided the foundation for a large body of work establishing that the response took place in the gonadotroph, that it was mediated by the GnRH receptor, and that it developed following puberty. We also found that the response is less than 5% as robust in males as in females and that expression of the gene was observed in vivo only during ovulation. Dentin matrix protein-1 had not previously appeared in the neuroendocrine literature, underscoring the potential impact that application of RNA sequencing can have on our understanding of biological mechanisms.
We stimulated the use of RNA sequencing within NIH through the establishment of an RNA interest group; it brings together a broad range of NIH scientists to discuss each other's work and to evaluate new technologies related to RNA sequencing.
Additional Funding
- United States–Israel Binational Science Foundation grant numbers 2009290 and 2005280
Publications
- Hartley SW, Coon SL, Savastano LE, Mullikin JC, NISC Comparative Sequencing Program, Fu C, Klein DC. Neurotranscriptomics: the effects of neonatal stimulus deprivation on the rat pineal transcriptome. PLoS One 2015; 10:e0137548.
- Falcón J, Coon SL, Besseau L, Cazaméa-Catalan D, Fuentès M, Magnanou E, Paulin C-H, Boeuf G, Sauzet S, Jorgensen EH, Mazan S, Wolf Y, Koonin EV, Steinbach PJ, Hyodo S, Klein, DC. Drastic neofunctionalization associated with evolution of the “timezyme” AANAT 500 million years ago. Proc Natl Acad Sci USA 2014; 111:314-319.
- Yamazaki F, Kim HH, Lau P, Hwang CK, Iuvone PM, Klein D, Clokie SJ. pY RNA1-s2: a highly retina-enriched small RNA that selectively binds to Matrin 3 (Matr3). PLoS One 2014; 9:e88217.
- Yamazaki F, Møller M, Fu C, Clokie SJ, Zykovich A, Coon SL, Klein DC, Rath MF. The Lhx9 homeobox gene controls pineal gland development and prevents postnatal hydrocephalus. Brain Struct Funct 2015; 220(3):1497-1509.
- Klein DC. Pineal gland and melatonin in endocrinology. Endocrinology: Adult and Pediatric 7th Edition. Eds: Jameson JL et al., Elsevier, St. Louis 2015; 312-322.
Collaborators
- Greti Aguilera, PhD, Program in Developmental Endocrinology and Genetics, NICHD, Bethesda, MD
- Peter Backlund, PhD, Biomedical Mass Spectrometry Core Facility, NICHD, Bethesda, MD
- Marjan Bozinoski, BA, Weill Cornell Medical College, New York, NY
- David Carter, PhD, University of Wales, Cardiff, United Kingdom
- Jack Falcón, PhD, Observeratoire Oceanologique/CNRS, Banyuls-sur-Mer, France
- Yoav Gothilf, PhD, Tel Aviv University, Tel Aviv, Israel
- Stephen Hartley, PhD, Genome Technology Branch, NHGRI, Rockville, MD
- P. Michael Iuvone, PhD, Emory University School of Medicine, Atlanta, GA
- Eugene Koonin, PhD, National Center for Biotechnology Information, NLM, Bethesda, MD
- Christopher E. Mason, PhD, Weill Cornell Medical College, New York, NY
- Morten Møller, MD, PhD, Panum Institute, University of Copenhagen, Copenhagen, Denmark
- Jim Mullikin, PhD, Genome Technology Branch, NHGRI, Rockville, MD
- Martin F. Rath, PhD, Panum Institute, Copenhagen University, Copenhagen, Denmark
- Leming Shi, PhD, Fudan University, Shanghai, China
Contact
For more information, email kleind@mail.nih.gov or visit http://sne.nichd.nih.gov.


