Interplay between Membrane Organelles, Cytoskeleton, and Metabolism in Cell Organization and Function

- Jennifer Lippincott-Schwartz, PhD, Head, Section on Organelle Biology
- Sarah Cohen, PhD, Visiting Fellow
- Uri Manor, PhD, Postdoctoral Fellow
- Carolyn Ott, PhD, Postdoctoral Fellow
- Christopher Obara, PhD, Postdoctoral Fellow
- Timothy Petri, PhD, Postdoctoral Fellow
- Prabuddha Sengupta, PhD, Postdoctoral Fellow
- Arnold Seo, PhD, Postdoctoral Fellow
- Alex Valm, PhD, Postdoctoral Fellow
- Aubrey Weigel, PhD, Postdoctoral Fellow
- Lingfeng Chen, PhD, Volunteer
- Prasanna Satpute, PhD, Volunteer
- Alex Ritter, BA, Graduate Student
- Bennett Waxse, BA, Graduate student
We investigate the global principles underlying cell behavior at both small and large spatial scales. At the small scale, we employ the super-resolution imaging techniques of photoactivated localization microscopy (PALM), interferometric 3D PALM, single-particle tracking PALM, and pair-correlation PALM to map the spatial organization, stoichiometry, and dynamics of proteins associated with various membrane-bound compartments and with the cytoskeleton. We also employ fluorescence photobleaching, photoactivation, fluorescence correlation, and fluorescence energy transfer methods to measure protein-protein interactions, protein turnover rates, and protein association rates. Such approaches allow us to assay cellular functions, including receptor stoichiometry and protein clustering and diffusion behavior at the nanometric scale in living cells. At the large scale, we investigate how complex behaviors of cells arise, such as cell crawling, polarization, cytokinesis, and viral budding. We study these complex behaviors by quantitatively analyzing diverse intracellular processes, including membrane trafficking, autophagy, actin/microtubule dynamics, and organelle assembly/disassembly pathways, which undergo dramatic changes as cells alter their behavior and organization throughout life. To assist these efforts, we combine various fluorescence-based imaging approaches, including total internal reflection fluorescence (TIRF) microscopy imaging and spinning-disk and laser-scanning confocal microscopy, with FRAP (fluorescence recovery after photobleaching), FLIP (fluorescence loss in photobleaching), and photoactivation to obtain large image data sets. We process the data sets computationally to extract biochemical and biophysical parameters, which can be related to the results of conventional biochemical assays. We then use the results to generate mechanistic understanding and predictive models of the behavior of cells and subcellular structures (including endoplasmic reticulum, Golgi, cilia, endosomes, lysosomes, autophagosomes, and mitochondria) under healthy and pathological conditions.
Click image to enlarge.
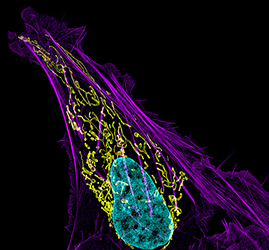
Figure 1. Mitochondrial and actin organization in a crawling cell
Structured illumination microscope (SIM) image of a U2OS (osteoblastoma) cell showing mitochondria stained with MITO-FRP (pink), actin stained with Alexa Fluor 488® phalloidin (green), and DNA stained with Hoechst 33342 (blue)
Actomyosin organization and its diverse functions
Cytotoxic T lymphocytes (CTLs) kill target cells by secreting granules containing perforin and granzymes into the immunological synapse (the site of contact formed between the CTL and target cell). We used spinning disk confocal and lattice light sheet microscopy to obtain unprecedented spatial and temporal resolution of the actin cytoskeleton and its role in both facilitating and limiting CTL secretion. We saw dynamic lamellapodial projections and a rearward flow of actin in migrating CTLs as they engaged a target cell. The synapse then formed in two stages: concentration of T cell receptors (TCRs) in the plasma membrane (PM) through lateral translocation (1 minute), followed by vesicular delivery of intracellular TCRs as the centrosome reached the synapse (6 minutes). Prior to synapse formation, a continuous actin meshwork underlies the entire PM; however, local clearing occurred as both the centrosome and granules docked. After several vesicles fused, the actin meshwork reappeared and secretion ceased. Actin clearance and reappearance correlated with the loss and gain of PtdIns(4,5)P2 (phosphatidylinositol 4,5-bisphosphate) in the contact zone. We concluded that the CTL contact zone is like a radially symmetric leading edge, with the distal region of protrusive actin polymerization being analogous to the lamellipodium, and the more central region, enriched in integrins and myosin IIA, analogous to the lamellum. The spatial-temporal regulation of actin in the contact zone serves to coordinate TCR docking and the timing of granule secretion.
In a second project, we examined the role of the actin cytoskeleton in regulating overall motion within the cytoplasm. We reasoned that ensemble forces from actomyosin activity could have a large effect on global motion within the cytoplasm, making these forces a critical readout of the dynamic state of the cell. To measure these forces and test how they control the motion of cytoplasmic components, we collaborated with physicist David Weitz, who devised a new methodology called force-spectrum-microscopy (FSM) to quantify force fluctuations within the cytoplasm. The technique combines measurements of the random motion of probe particles with independent micro-mechanical measurements of the cytoplasm. Increased cytoplasmic force fluctuations substantially enhanced intracellular movement of small and large components, including organelles. Cytoplasmic force fluctuations varied between cell types and were three times larger in malignant cells than in normal cells. Separately, in close collaboration with Christoph Schmidt, we found that force generation by the actin cytoskeleton surrounding the basal body causes previously undocumented active primary cilia movements, which could be important for tuning and calibrating ciliary sensory functions. The results of these studies reveal that actomyosin dynamics are a critical readout of proper cell health and have major effects on diverse cellular functions.
Click image to enlarge.
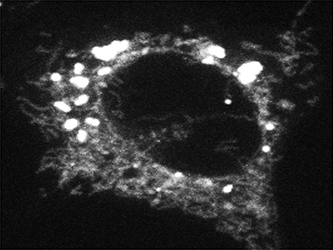
Figure 2. Fatty acid trafficking from lipid droplets to mitochondria
Confocal microscopy of a cell whose lipid droplets were labeled with a fluorescent fatty acid probe and then starved to visualize movement of the fatty acid from lipid droplets to mitochondria
Mitochondrial dynamics
To study mammalian cell adaptation to nutrient starvation, we examined the interplay between mitochondrial fusion dynamics, autophagy, fatty acid (FA) trafficking, and lipid droplets (LDs). Given that cells appear to adapt to nutrient starvation by shifting their metabolism from reliance on glucose metabolism to utilization of mitochondrial FA oxidation, we developed an assay to investigate how FAs become mobilized and delivered to mitochondria. Using a pulse-chase labeling method to visualize movement of FAs in live cells, we demonstrated that starved cells primarily use LDs as a conduit to supply mitochondria with FAs for β-oxidation. This occurred through lipase-mediated FA mobilization from mitochondria-associated LDs, rather than autophagy (contrary to the pathway used by yeast cells). Autophagy contributed to the altered metabolic scheme by recovering lipids from degraded organelles, which could be used to refill LDs. Notably, mitochondrial tubulation was essential for distribution of FAs throughout the mitochondrial network. Defects in mitochondrial fusion led to massive alterations in cellular FA routing. Not only were non-metabolized FAs redirected to and stored in LDs, they were excessively expelled from cells. Given that FAs are toxic at high levels and serve as signaling molecules at low levels, the results suggest that defects in mitochondrial dynamics and FA trafficking pathways may underlie the pathologies of many metabolic diseases such as diabetes and obesity.
In a separate mitochondria-related project, we uncovered new machinery regulating mitochondrial fission. Seminal work from others showed that, before mitochondrial division by the dynamin-related protein Drp1, endoplasmic reticulum (ER) tubules encircle and constrict mitochondria. Constriction results from actin polymerization controlled by the ER–localized formin protein INF2. However, how ER tubules recognize mitochondria and facilitate fission is unclear. In investigating this question, we discovered a novel mitochondria-localized actin-nucleating protein, Spire1C, which interacts with INF2 on the ER. Cooperation between Spire1C and INF2 enhanced actin assembly selectively at ER/mitochondria intersections, facilitating mitochondrial constriction. We are proposing, therefore, that, during mitochondrial division, a Spire1C–INF2 interaction tethers the ER to mitochondria and mediates actin polymerization, resulting in mitochondrial constriction.
Additional Funding
- Pharmacology Research Associate Training Program
Publications
- Manor U, Bartholomew S, Golani G, Christenson E, Kozlov M, Higgs H, Spudich J, Lippincott-Schwartz J. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. eLife 2015; 4:08828.
- Ritter AT, Asano Y, Stinchcombe JC, Dieckmann NM, Chen BC, Gawden-Bone C, van Engelenburg S, Legant W, Gao L, Davidson MW, Betzig E, Lippincott-Schwartz J, Griffiths GM. Actin depletion initiates events leading to granule secretion at the immunological synapse. Immunity 2015; 42(5):864-876.
- Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell 2015; 32:678-692.
- Battle C, Ott CM, Burnette DT, Lippincott-Schwartz J, Schmidt CF. Intracellular and extracellular forces drive primary cilia movement. Proc Natl Acad Sci USA 2015; 112(5):1410-1415.
- Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, Liu B, Chang C, Zhou T, Lippincott-Schwartz J, Liu W. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell 2015; 57(3):456-466.
Collaborators
- Eric Betzig, PhD, Howard Hughes Medical Institute, Janelia Farm Research Campus, Ashburn, VA
- Hu Cang, PhD, Waitt Advanced Biophotonics Center, Salk Institute for Biological Studies, La Jolla, CA
- Eric O. Freed, PhD, HIV Drug Resistance Program, Center for Cancer Research, NCI, Frederick, MD
- Dong Fu, PhD, Cell Biology and Metabolism Program, NICHD, Bethesda, MD
- Gillian M. Griffiths, PhD, FMedSci, FRS, University of Cambridge and Cambridge Institute for Medical Research, Cambridge, UK
- Wei Liu, MD, PhD, Zhejiang University School of Medicine, Hangzhou, China
- Mark Marsh, PhD, University College London, London, United Kingdom
- Christoph Schmidt, PhD, Georg-August-Universität, Göttingen, Germany
- David A. Weitz, PhD, Harvard University School of Engineering and Applied Sciences, Cambridge, MA
- Christopher Westlake, PhD, Laboratory of Cell and Developmental Signaling, Center for Cancer Research, NCI, Frederick, MD
- Xuebiao Yao, PhD, Hefei National Laboratory for Physical Sciences at the Nanoscale and University of Science and Technology of China, Hefei, China
Contact
For more information, email lippincj@mail.nih.gov or visit http://lippincottschwartzlab.nichd.nih.gov.


