Transcriptional and Translational Regulatory Mechanisms in Nutrient Control of Gene Expression

- Alan G. Hinnebusch, PhD, Head, Section on Nutrient Control of Gene Expression
- Hongfang Qiu, PhD, Staff Scientist
- Neelam Sen, PhD, Research Fellow
- Suna Gulay, PhD, Postdoctoral Fellow
- Anil Thakur, PhD, Postdoctoral Fellow
- Jyothsna Visweswaraiah, PhD, Postdoctoral Fellow
- Fnu Yashpal, PhD, Postdoctoral Fellow
- David Young, PhD, Postdoctoral Fellow
- Quira Zeiden, PhD, Postdoctoral Fellow
- Fujun Zhou, PhD, Postdoctoral Fellow
- Jinsheng Dong, PhD, Senior Research Assistant
- Fan Zhang, MS, Senior Research Assistant
- Cuihua Hu, BA, Research Assistant
- Laura Marler, BS, Graduate Student
We study molecular mechanisms of gene regulation at the translational and transcriptional levels, using the regulation of amino acid–biosynthetic genes in budding yeast as a model system. Transcription of these and many other genes is coordinately induced by the transcriptional activator Gcn4 in response to starvation of any amino acid. Expression of GCN4 is coupled to amino acid levels by a conserved translational control mechanism involving upstream open reading frames (uORFs) in GCN4 mRNA. Ribosomes translate the 5′-most uORF (uORF1) and, under non-starvation conditions, reinitiate translation at uORFs 2, 3, or 4 and then dissociate from the mRNA, keeping GCN4 translation repressed. In starvation conditions, the reinitiating ribosomes bypass uORFs 2–4 and reinitiate at GCN4 instead, owing to lowered availability of the ternary complex (TC)—comprised of initiation factor 2 (eIF2), GTP, and initiator Met-tRNAi—which binds to the small (40S) ribosomal subunit to assemble a 43S preinitiation complex (PIC). TC abundance is reduced in starved cells by phosphorylation of the alpha subunit of eIF2 (eIF2α) by Gcn2, a protein kinase conserved in all eukaryotes, converting eIF2 from substrate to inhibitor of its guanine nucleotide exchange factor (GEF) eIF2B. Hence, GCN4 translation is an in vivo indicator of impaired TC loading on 40S subunits. We previously exploited this fact to isolate mutations in subunits of eIF2B that constitutively derepress GCN4 (Gcd− phenotype) by lowering TC assembly in the absence of eIF2 phosphorylation. More recently, we used the Gcd− selection to identify domains/residues in eIF1, eIF1A, and eIF3, and also residues of 18S rRNA located near the 'P' decoding site of the 40S subunit, that participate in rapid TC recruitment in vivo. In collaboration with Jon Lorsch's group, we demonstrated that segments/residues in eIF1, eIF1A, and 18S rRNA, which are implicated genetically in TC recruitment, also stimulate this reaction in a fully reconstituted in vitro system.
We also investigate the roles of various eIFs and the 40S subunit in scanning the mRNA 5′ untranslated region and in accurately identifying the AUG initiation codon. The studies exploit a genetic selection for mutations that elevate initiation at near-cognate UUG start codons (Sui− phenotype) or suppress this aberrant initiation event (Ssu− phenotype). In this way, we showed that eIF1A and the c-subunit of eIF3 make critical contributions to accurate AUG recognition, and localized these functions to the unstructured N- and C-tails of eIF1A and the N-terminal domain of eIF3c. Also in collaboration with Lorsch's group, we provided strong evidence that dissociation of eIF1 from the PIC is a critical step in AUG recognition, indicating that eIF1 serves as a 'gate-keeper' necessary to suppress initiation at non-AUG triplets. Biochemical analysis revealed that eIF1 stabilizes an 'open' conformation of the 40S subunit conducive to scanning and loading of TC in a metastable state[P(OUT)], which enables inspection of successive mRNA triplets in the P site for complementarity with the anticodon of Met-tRNAi. However, eIF1 must dissociate from the PIC for AUG selection, as the absence of eIF1 favors the closed, scanning-arrested conformation of the 40S subunit with TC more tightly bound [P(IN) state]. We recently established that basic residues in eIF1's helix-1 and β-hairpin-1 mediate 40S binding and stabilize the open, P(OUT) complex to promote Met-tRNAi recruitment and accurate AUG selection in vivo. Previously, we showed that 10–amino acid repeats in the C-terminal tail (CTT) of eIF1A (dubbed "scanning enhancers" or SE elements) function together with eIF1 to stabilize the open, P(OUT) complex but must be antagonized by scanning-inhibitory (SI) elements in the N-terminal tail (NTT) and helical domain of eIF1A to achieve rearrangement to the closed, P(IN) complex and start codon recognition. In collaboration with Lorsch's group, we recently established that the eIF1A CTT moves towards the N-terminal domain (NTD) of eIF5 in response to AUG recognition, and that this movement—which depends on the eIF1A SE elements—is required for Pi release from eIF2. We proposed that accommodation of tRNAi in the P(IN) state triggers eIF1 displacement, which in turn triggers movement of the eIF1A-CTT and Pi release.
The eIF4F complex, which binds to the mRNA 5′ cap structure, is thought to resolve mRNA secondary structure to enhance recruitment of the 43S PIC and scanning of the mRNA leader. We recently dissected the NTD of eIF4G, the scaffolding subunit of eIF4F—harboring binding sites for cap-binding factor eIF4E, poly(A)-tail-binding factor PABP, and RNA helicase eIF4A—and identified conserved elements (RNA1, boxes 1–3) that functionally overlap to promote eIF4G interaction with mRNA and PABP, thereby activating mRNA for PIC attachment. We also identified a novel role for cofactor eIF4B in promoting eIF4AV/eIF4G association (eIF4F assembly) in vivo and, in collaboration with Lorsch's group, showed that binding of eIF4B near the mRNA entry channel of the 40S subunit is likely to be crucial for stimulating eIF4F function in PIC recruitment.
In the arena of transcriptional control, we previously defined multiple clusters of hydrophobic residues that constitute the transcriptional activation domain of Gcn4, identified co-activators required for gene activation by Gcn4 in vivo, and defined the molecular program for recruitment of nucleosome-modifying and -remodeling enzymes and adaptor proteins that remove repressive chromatin structure and recruit TATA–binding protein and RNA polymerase II (Pol II) to Gcn4 target promoters. We also demonstrated co-transcriptional recruitment of the histone acetyltransferase (HAT) complexes SAGA and NuA4 to transcribed coding sequences and showed that they cooperate to enhance transcription-associated histone eviction and Pol II elongation in vivo. We described a two-stage recruitment mechanism for NuA4, involving the serine-5–phosphorylated C-terminal domain (CTD) of Pol II and methylated histones, and more recently we extended this mechanism to include the histone deacetylase (HDA) complex Rpd3C(S). In addition, we showed that the HDAs Rpd3, Hos2, and Hda1 have overlapping functions in deacetylating coding-sequence nucleosomes and obtained evidence that histone acetylation is a key determinant of co-transcriptional nucleosome disassembly.
More recently, we elucidated the roles of the Pol II CTD kinases Kin28 and Bur1 and the C-terminal repeats (CTRs) of the Spt5 subunit of DSIF in recruitment of transcription elongation factor Paf1C. We showed that Ser5-CTD phosphorylation by Kin28 enhances recruitment of Bur1 near promoter regions where Bur1 contributes to Ser2–CTD phosphorylation. We then established that Kin28 enhances Paf1C recruitment by: (1) promoting Bur1 recruitment via the phosphorylated Pol II CTD, with attendant phosphorylation of Spt5 CTRs by Bur1; and (2) collaborating with Bur1 to generate Ser2–/Ser5–diphosphorylated Pol II CTD repeats. The resulting phosphorylated CTD and CTR repeats bind to distinct Paf1C subunits to enable efficient recruitment of the entire complex to elongating Pol II.
Biochemical evidence for conformational changes in the P site and mRNA entry channel evoked by AUG recognition in reconstituted PICs
It is thought that the scanning PIC assumes an open conformation and that AUG recognition evokes a closed state that arrests scanning with more stable Met-tRNAi binding [P(IN) state], with attendant displacement of the eIF1A-CTT from the P site and dissociation of eIF1 from the 40S subunit. We obtained physical evidence for these conformational rearrangements by comparing patterns of directed hydroxyl radical cleavage (DHRC) of rRNA by Fe(II)-BABE tethered to unique cysteine residues engineered in eIF1A, in PICs reconstituted with mRNA with AUG or near-cognate (AUC) start codons. Key rRNA residues in the P site displayed reduced cleavage in AUG versus AUC PICs, suggesting that accessibility of these rRNA residues is reduced by their increased interaction with Met-tRNAi in the P(IN) state. The cleavage data also indicate that AUG recognition evokes dissociation of eIF1 from its 40S binding site, a constricted conformation of the entry channel, and displacement of the eIF1A-CTT from near the P-site.
Structures of yeast preinitiation complexes reveal conformational changes from mRNA scanning to start-codon recognition.
In colloboration with Venki Ramakrishnan's lab and Jon Lorsch's group, we obtained cryo-EM reconstructions of yeast PICs that represent different stages of the initiation pathway, at 3.5–6.1 Å resolution. These include 40S-eIF1-eIF1A complexes and partial yeast 48S PICs in open and closed conformations (py48S-open and py48-closed). The structures provide a wealth of new information about conformational changes occurring in the transition from scanning to AUG selection. Comparing the 40S-eIF1-eIF1A complex with the free 40S reveals rotation of the 40S head that might promote TC binding to form the 43S PIC. The py48S-open, formed using mRNA with AUC start codon, reveals an upward shift of the 40S head that widens the mRNA entry channel and opens its latch—consistent with our eIF1A/Fe(II)-BABE cleavage data—which should facilitate mRNA insertion into the binding cleft to form the scanning PIC. Moreover, the P site is widened and lacks tRNAi contacts with the 40S body present in canonical 80S-tRNAi complexes. By contrast, py48S-closed, formed with mRNA(AUG), reveals downward head movement that closes the latch, clamps the mRNA into the binding cleft, and fully encloses tRNAi in the P site. The eIF1A NTT assumes a structured conformation and interacts with the AUG:anticodon duplex, consistent with its role in stabilizing P(IN). eIF1 is repositioned on the 40S and deformed to prevent a clash with tRNAi, likely as a prelude to eIF1 dissociation from the 40S subunit. Both py48S-open and -closed complexes reveal eIF2beta and portions of the eIF3 complex. The eIF3 trimeric subcomplex eIF3b-CTD/eIF3i/eIF3g-NTD resides on the subunit-interface surface of the 40S and appears to lock mRNA into the 40S binding cleft. eIF2beta interacts with tRNAi and segments of eIF1 and eIF1A exclusively in py48S-open, which could stabilize binding of TC and eIF1 to the scanning PIC prior to AUG recognition. A portion of eIF2alpha domain-I projects into the entry channel and contacts the beta-hairpin of uS7/Rps5 and nucleotides just upstream of the AUG codon, including the key –3 nucleotide of the Kozak consensus sequence for efficient AUG selection, consistent with a proposed role for eIF2alpha in start codon recognition.
Exit-channel beta-hairpin of 40S ribosomal protein Rps5/u7 is a critical determinant of efficient and accurate translation initiation.
In the py48S complexes described above, the beta-hairpin of 40S protein Rps5/uS7 protrudes into the mRNA exit-channel, contacting the TC and mRNA context nucleotides; however, its importance in AUG selection was unknown. We identified substitutions in beta-strand 1 (E144R) and a nearby C-terminal residue (R225K) of yeast Rps5 that reduced bulk initiation, conferred leaky scanning of an upstream AUG, and lowered initiation fidelity by exacerbating the effect of poor context of the eIF1 start codon to thereby reduce eIF1 abundance. After reinstating WT eIF1 abundance with an extra copy of its gene (SUI1), both Rps5 substitutions were found to suppress UUG initiation, consistent with their discrimination against the poor-context SUI1 AUG codon. Consistent with this, in vitro analysis of mutant ribosomes showed that E144R greatly destabilized P(IN), increasing the dissociation rate (koff) of TC from reconstituted 43S-mRNA PICs with AUG or UUG start codons. Other substitutions in beta-hairpin loop residues also elevated initiation fidelity, suppressing UUG initiation in cells expressing Sui– variant eIF2beta-S264Y; one such mutant (R148E) destabilized P-in at UUG, but not AUG, start codons in PICs reconstituted with the eIF2beta-S264Y form of eIF2. Thus, the Rps5 beta-hairpin is crucial for efficient and accurate start codon recognition in vivo.
Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A.
RNA helicases eIF4A and Ded1 are believed to resolve mRNA secondary structures that impede ribosome attachment to the mRNA or subsequent scanning to the start codon, but whether they perform unique or overlapping functions in vivo is poorly understood. We compared the effects of mutations in Ded1 or eIF4A on global translational efficiencies (TEs) in yeast by ribosome footprint profiling. Despite similar reductions in bulk translation, inactivation of a cold-sensitive Ded1 mutant substantially reduced the TEs of over 600 mRNAs, whereas inactivation of a temperature-sensitive eIF4A mutant yielded fewer than 40 similarly impaired mRNAs. Ded1–dependent mRNAs exhibit greater than average 5′ UTR length and propensity for secondary structure, implicating Ded1 in scanning though structured 5′ UTRs. Reporter assays confirmed that cap-distal stem-loop insertions raised dependence on Ded1 but not on eIF4A for efficient translation. Our findings suggest that Ded1 is critical to promote scanning through secondary structures in 5′ UTRs, while eIF4A promotes a step of initiation common to virtually all yeast mRNAs.
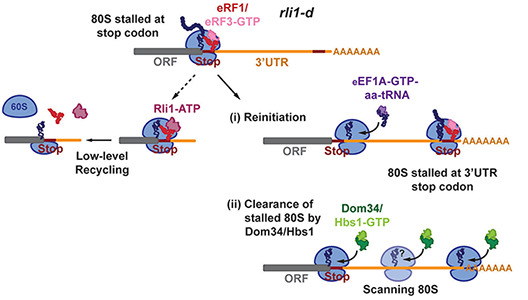
Rli1/ABCE1 recycles terminating ribosomes and controls reinitiation in 3′ UTRs in vivo.
Ribosome recycling occurs after release of completed polypeptides at stop codons, and involves dissociation of the 60S subunit from the 80S post-termination complex (post-TC). Rli1/ABCE1 (translation initiation factor/ATP-binding cassette E1) catalyzes this reaction in vitro, but it was unknown whether Rli1 is essential for recycling in vivo. Ribosome profiling of yeast cells depleted of Rli1 revealed accumulation of 80S ribosomes at the stop codons and adjoining 3′ UTRs of most genes, consistent with defective recycling that allows ribosomes to proceed into 3′ UTRs. The distribution of footprints indicates that 3′ UTR ribosomes are translating, with footprint peaks at 3′ UTR stop codons in all three frames, which are generally diminished by in-frame upstream stop codons and exhibit the atypical long fragment sizes characteristic of ribosomes at canonical stop codons. Moreover, histidine starvation evokes footprint peaks at 3′ UTR histidine codons upstream from prominent stop codon–stall sites in the same reading frame. Novel 3′ UTR translation products were detected by western analysis after inserting epitope tags immediately before stop codon-stall sites. The small sizes of the tagged 3′ UTR translation products, combined with their sequences determined by mass spectrometry, indicate that un-recycled post-TCs migrate a short distance from the stop codon and reinitiate translation without apparent codon preference. In a double mutant lacking also the 80S rescue factor Dom34, Rli1 evokes a dramatic increase in 3′ UTR ribosomes at densities comparable to the upstream coding sequences, indicating that Dom34 is responsible for rescuing unrecycled 80S ribosomes. Overexpressing Dom34 in wild-type eliminates the low-level 80S occupancy in 3′ UTRs and the expression of a tagged 3′ UTR reinitiation product, indicating that native Rli1 is insufficient to prevent all 3′ UTR translation by post-TCs in normal cells. Thus, Rli1 is required for ribosome recycling in vivo and acts with Dom34 to prevent aberrant reinitiation and translation of 3′ UTRs (see model in Figure).
Click image to enlarge.
Schematic model depicting the fate of post-termination ribosomal complexes (post-TCs) on depletion of Rli1/ABCE1 in an rli1-d degron mutant
Recognition of the stop codon of the main open-reading frame (ORF) at each gene by translation termination factors eRF1/eRF3-GTP (top row) is followed by release of the completed polypeptide and dissociation of eRF3-GDP (not depicted). Any residual Rli1 remaining in the rli1-d cells could bind to post-TCs (ternary complexes) and catalyze dissociation of the 60S subunit (middle row, left). However, many post-TCs are not recycled owing to Rli1 deficiency, migrate a short distance from the stop codon, reinitiate translation, and frequently terminate at a 3′ UTR stop codon to produce a 3′ UTR–encoded polypeptide (middle row, right). Such reinitiation events appear to be diminished by ribosome rescue factor Dom34, potentially because post-TC ribosomes are rescued at the main ORF stop codons or as they begin scanning. Any ribosomes that reach the 3′ UTR/poly(A) boundary by reinitiation or scanning are also rescued by Dom34.
Analysis of factors mediating nucleosome disassembly at Gcn4 target gene promoters in vivo
A key unsolved aspect of transcriptional activation by Gcn4 is how it mediates the eviction of nucleosomes that occlude promoter DNA sequences and block access by GTFs (general transcription factors) and Pol II. Indeed, the mechanism of this key step of gene activation and the impact of defective nucleosome eviction on transcription are not fully understood for any yeast genes. Previous studies implicated histone chaperones, chromatin remodelers, or histone acetyltransferases in the remodeling or eviction of nucleosomes from the promoters of certain yeast genes, but it was unclear whether these co-factors function broadly in nucleosome eviction, given that co-factor requirements at most yeast promoters are unknown. Eviction of promoter nucleosomes is considered to be rate-limiting for transcriptional activation, but the consequences of impaired nucleosome eviction on transcription have not been analyzed genome-wide. We addressed these questions by analyzing histone H3 eviction for the hundreds of genes in the Gcn4 transcriptome on induction of Gcn4 in a large panel of mutants lacking one or more co-factors implicated at particular yeast genes. By conventional chromatin immunoprecipitation analysis (ChIP) of four canonical Gcn4 target genes, ARG1, HIS4, ARG4, and CPA2, we excluded a requirement for several co-factors implicated previously at other genes (e.g., Asf1, Nap1, RSC) and implicated the remodeler SWI/SNF (Snf2), HAT Gcn5, and Hsp70 co-chaperone Ydj1 in nucleosome eviction at these Gcn4 target genes. Expanding our analysis genome-wide by H3 ChIP-Seq, we found that Snf2, Gcn5, and Ydj1 collaborate in evicting H3 from the −1 and +1 promoter nucleosomes and from the intervening nucleosome-depleted region (NDR) at a large fraction of the Gcn4 transcriptome. The three cofactors were found to function similarly at virtually all yeast promoters. Surprisingly, however, defective H3 eviction in co-factor mutants was coupled with reduced transcription (Pol II densities measured by Rpb3 ChIP-Seq) for a subset of genes only, which included the induced Gcn4 transcriptome and the most highly expressed subset of constitutively expressed yeast genes. In fact, the most weakly expressed genes displayed an increase in transcription relative to other genes in response to global attenuation of nucleosome eviction. Thus, we established that steady-state eviction of promoter nucleosomes is required for maximal transcription of highly expressed genes, and that Gcn5, Snf2, and Ydj1 function broadly in this step of gene activation, but discovered unexpectedly that some other aspect of transcriptional activation is more generally rate-limiting for transcription of most genes in amino acid–deprived yeast.
Publications
- Zhang F, Saini AK, Shin BS, Nanda J, Hinnebusch AG. Conformational changes in the P site and mRNA entry channel evoked by AUG recognition in yeast translation preinitiation complexes. Nucleic Acids Res 2015; 43:2293-2312.
- Sen ND, Zhou F, Ingolia NT, Hinnebusch AG. Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res 2015; 25:1196-1220.
- Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, Green R. Rli1/ABCE1 recycles terminating ribosomes and controls reinitiation in 3’UTRs in vivo. Cell 2015; 162:872-884.
- Llacer JL, Hussain T, Marler L, Aitken CE, Thakur A, Lorsch JR, Hinnebusch AG. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell 2015; 59:399-412.
- Visweswaraiah J, Pittman Y, Dever TE, Hinnebusch AG. The ß-hairpin of 40S exit channel protein Rps5/uS7 promotes efficient and accurate translation initiation in vivo. eLife 2015; 4:e07939.
Collaborators
- Katsura Asano, PhD, Kansas State University, Manhattan, KS
- David Clark, PhD, Program in Genomics of Differentiation, NICHD, Bethesda, MD
- Rachel Green, PhD, The Johns Hopkins University School of Medicine, Baltimore, MD
- Nicholas Ingolia, PhD, University of California Berkeley, Berkeley, CA
- Jon Lorsch, PhD, The Johns Hopkins University School of Medicine, Baltimore, MD
- Venkatraman Ramakrishnan, PhD, MRC Laboratory of Molecular Biology, Cambridge, UK
- Gerhard Wagner, PhD, Harvard Medical School, Boston, MA
Contact
For more information, email hinnebua@mail.nih.gov or visit http://sncge.nichd.nih.gov.


