Epigenome Reprogramming During Mammalian Development

- Todd S. Macfarlan, PhD, Head, Unit on Mammalian Epigenome Reprogramming
- Carson Miller, PhD, Postdoctoral Fellow
- Gernot Wolf, PhD, Visiting Fellow
- Peng Yang, PhD, Visiting Fellow
- Don Hoang, BS, Postbaccalaureate Fellow
- Matthew Tinkham, BS, Postbaccalaureate Fellow
- Sherry Ralls, BA, Biologist
- Justin Demmerle, AB, Graduate Student
Embryonic development requires the coordinated activities of transcription factors that bind to DNA and the enzymatic machinery that alters the chromatin state of DNA to “dial” gene expression programs and achieve a prodigious diversity of transcriptional outputs. This includes the activation of genes necessary to carry out cellular functions in distinct cell types, but also suppression of parasitic mobile elements, including endogenous retroviruses (ERVs), that are a threat to genomic stability. Our objective is to understand the dichotomy between transcription factors and chromatin. On the one hand, transcription factors bind to DNA to guide the formation of specific heritable chromatin states (which we explore by studying the initiation of heterochromatin formation by the KRAB-ZFP protein family), but on the other, the chromatin state also imposes a layer of control on how transcription factors interact with DNA, effectively “gating” transcription factor occupancy (which we explore by studying the effect of chromatin state on transcription factors implicated in motor neuron development). The tools we use to explore these questions are the same. We apply cutting-edge genetic and synthetic biology approaches to mouse embryonic stem cell–based models of development, allowing us to perform biochemical and next-generation sequencing experiments, which require large cell numbers. These in vitro approaches complement in vivo studies with mutant and transgenic mice to give us a unique mechanistic understanding of transcriptional regulation in mammalian development. Such foundational studies will lay the framework for our long-term objectives: to be able to use transcription factors in a rational way for cellular programming such that we can build better models of human disease and open new avenues in regenerative medicine.
Regulation of endogenous retroviruses by KRAB-zinc finger proteins
Retroviruses pose a threat to human health by infecting somatic cells, but retroviruses have also been infecting our mammalian ancestors for millions of years, accumulating in the germ-line as ERVs that account for nearly 10% of our genomic DNA. The laboratory studies ERVs from two perspectives: (1) as parasites that must be kept in check by the host to prevent widespread viral activation; and 2) as symbionts that can be co-opted by the host for evolutionary advantage. Our objective is to understand how the host has adapted recognition machinery to establish stable epigenetic silencing of ERVs, how ERVs sometimes evade these silencing mechanisms, and how the evasive activities have led to host co-option of viral regulatory sequences that may have contributed to evolution of mammals. We hypothesize that the rapidly diversifying KRAB-ZFP family plays a critical role in the recognition and silencing of ERVs.
Krüppel-associated box zinc finger proteins (KRAB-ZFPs) have emerged as candidates that recognize ERVs. KRAB-ZFPs are rapidly evolving transcriptional repressors that emerged in tetrapods. They make up the largest family of transcription factors in mammals (estimated to be about 200–300 in mice and humans)1. Each species has its own unique repertoire of KRAB-ZFPs, with a small number shared with closely related species and a larger fraction specific to each species. Despite their abundance, little is known about their physiological functions. KRAB-ZFPs consist of an N-terminal KRAB domain that binds to the co-repressor KAP12 and a variable number of C-terminal C2H2 zinc finger domains that mediate sequence-specific DNA binding. KAP1 directly interacts with the KRAB domain3, which recruits the histone methyltransferase (HMT) SETDB1 and heterochromatin protein 1 (HP1) to initiate heterochromatic silencing4,5. Several lines of evidence point to a role for the KRAB-ZFP family in ERV silencing. First, the number of C2H2 zinc finger genes in mammals correlates with the number of ERVs6. Second, the KRAB-ZFP protein ZFP809 was isolated based on its ability to bind to the primer binding site for proline tRNA (PBSPro) of murine leukemia virus (MuLV)7. Third, deletion of the KRAB-ZFP co-repressors Trim28 or Setdb1 leads to activation of many ERVs8,9. Thus, we have begun a systematic interrogation of KRAB-ZFP function as a potential adaptive repression system against ERVs.
We focused on ZFP809 as a likely ERV–suppressing KRAB-ZFP owing to the fact that it was originally identified as part of a repression complex that recognizes infectious MuLV via direct binding to the 18 nt PBSPro sequence7,10. We hypothesized that ZFP809 might function in vivo to repress other ERVs that utilized the PBSPro. Using ChIP-seq of epitope-tagged ZFP809 in embryonic stem cells (ESCs) and embryonic carcinoma (EC) cells, we determined that ZFP809 bound to several sub-classes of ERV elements via the PBSPro. We generated Zfp809 knockout (KO) mice to determine whether ZFP809 was required for the silencing of VL30Pro, an endogenous retroviral element that contains PBSPro. We found that Zfp809 KO tissues display high levels of VL30Pro elements and that the targeted elements display an epigenetic shift from repressive epigenetic marks (H3K9me3 and CpG methylation) to active marks (H3K9Ac and CpG hypo-methylation). ZFP809–mediated repression extended to a handful of genes that contained adjacent VL30Pro integrations. Furthermore, using a combination of conditional alleles and rescue experiments, we determined that ZFP809 activity is required in development to initiate silencing, but not in somatic cells to maintain silencing. The studies provided the first demonstration for the in vivo requirement of a KRAB-ZFP in the recognition and silencing of ERVs.
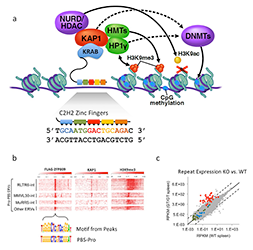
Click image to enlarge.
ZFP809 represses VL30Pro LTR-retroelements.
a. Model of KRAB-ZFP protein function. KRAB-ZFPs consist of KRAB–repression domain that interacts with KAP1 to assemble a corepressor complex, establishing stable epigenetic silencing of targets. Tandem C2H2 zinc finger (ZF) domains mediate sequence-specific DNA interactions, with each ZF interacting with three nucleotides of target DNA. b. Heat map plots of ChIP-seq data (with indicated antibodies) of Flag-ZFP809 peak regions. Peaks were centered on PBSPro–like motif and grouped according to repeat masker annotation. c. RNA-seq differential expression analysis of all repeat masker annotated elements in Zfp809 wild-type (WT) vs. knockout (KO) spleen. Red dots indicate VL30Pro elements, most of which are reactivated in KOs.
KRAB-ZFPs such as ZFP809 initiate ERV silencing by establishing the methylation of histone H3 on lysine 9 via the recruitment of the histone methylatransferase SETDB1. Mice have three histone H3 variants (H3.1, H3.2, and H3.3), and whether one or more of these variants was critical for ERV silencing had not been explored. Using ChIP-seq in primary mouse embryonic fibroblasts (PMEFs) and induced pluripotent stem cells (iPSCs) containing genetically tagged histone H3.3 genes, we found a strong enrichment of the variant histone H3.3 at ERVs co-occupied by KAP1, SETDB1, and H3K9me3, including VL30Pro elements recognized by ZFP809. Importantly, this enrichment was present only in pluripotent cells. We therefore explored the possibility that the deposition of histone H3.3 might be required for ERV silencing. To test this hypothesis, we used CRISPR/Cas9 to create a homozygous 'floxed' Daxx gene, which had previously been shown to be responsible for H3.3 deposition at telomeres in ESCs11. We found that acute loss of Daxx by Cre-mediated recombination in ESCs caused a complete loss of histone H3.3 at ERVs, but only very minor ERV reactivation. The data suggest that DAXX–dependent deposition of histone H3.3 is dispensable for ERV silencing. The finding is in conflict with a recent report that argued that some ERVs display reactivation in histone H3.3 KO ESCs, and that IAP elements, in particular, become transpositionally active12. Our re-analysis of this dataset challenges this central conclusion. We found that there is no correlation between ERVs marked by histone H3.3 and those showing reactivation in H3.3 KOs; furthermore, we demonstrated that the reported IAP 're-integrations' are not a result of retrotransposition, but were simply polymorphic IAP ERV elements mixed into the genetic background of the ESCs used for the study. Our data support a model in which histone H3.3 is deposited into ERVs in pluripotent cells, but that this deposition is not required for ERV silencing, which has been supported further in the recent literature13,14.
1Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol 2003;4:231.
2Friedman JR et al. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev 1996;10:2067-2078.
3Peng H et al. Biochemical analysis of the Kruppel-associated box (KRAB) transcriptional repression domain. J Biol Chem 2000;275:18000-18010.
4Ryan RF et al. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol 1999;19:4366-4378.
5Schultz DC et al. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 2002;16:919-932.
6Thomas JH, Schneider S. Coevolution of retroelements and tandem zinc finger genes. Genome Res 2011;21:1800-1812.
7Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 2009;458:1201-1204.
8Rowe HM et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 2010;463:237-240.
9Matsui T et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 2010;464:927-931.
10Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 2007;131:46-57.
11Goldberg AD et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010;140:678-691.
12Elsasser SJ et al. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 2015;522:240-244.
13Jang CW et al. Histone H3.3 maintains genome integrity during mammalian development. Genes Dev 2015;29:1377-1392.
14Sadic D et al. Atrx promotes heterochromatin formation at retrotransposons. EMBO Rep 2015;16:836-850.
Additional Funding
- Scientific Director's Award, 2014-2015, Epigenetic Memory that directs iPSC reprogramming: the role of H3.3
Publications
- Wolf G, Yang P, Füchtbauer A, Füchtbauer EM, Silva AM, Park C, Nielsen AL, Pedersen FS, Macfarlan TS. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev 2015; 29:538-554.
- Wang J, Telese F, Tan Y, Li W, Jin C, He X, Basnet H, Ma Q, Merkurjev D, Zhu X, Liu Z, Zhang J, Ohgi K, Taylor H, White RR, Macfarlan TS, Pfaff SL, Rosenfeld MG. LSD1n is a H4K20 demethylase regulating memory formation via transcriptional elongation control. Nat Neurosci 2015; 18:1256-1264.
- Lee H, Kim M, Kim N, Macfarlan T, Pfaff SL, Mastick GS, Song MR. Slit and Semaphorin signaling governed by Islet transcription factors positions motor neuron somata within the neural tube. Exp Neurol 2015; 269:17-27.
- Wolf G, Greenberg D, Macfarlan TS. Spotting the enemy within: targeted silencing of foreign DNA in mammalian genomes by the Kruppel-associated box zinc finger protein family. Mobile DNA 2015; 6:17.
- Wolf G, Macfarlan TS. Revealing the complexity of retroviral repression. Cell 2015; 163:30-32.
Collaborators
- Shigeke Iwase, PhD, University of Michigan Medical School, Ann Arbor, MI
- Keiko Ozato, PhD, Program in Genomics of Differentiation, NICHD, Bethesda, MD
- Finn Skou Pedersen, PhD, Aarhus Universitet, Aarhus, Denmark
- Michael G. Rosenfeld, MD, Howard Hughes Medical Institute, University of California San Diego, La Jolla, CA
- Mi-Ryoung Song, PhD, The Gwangju Institute of Science and Technology, Gwangju, South Korea
Contact
For more information, email todd.macfarlan@nih.gov or visit http://macfarlan.nichd.nih.gov.


