Chromosome Segregation in Higher Eukaryotes
- Mary Dasso, PhD, Head, Section on Cell Cycle Regulation
- Alexei Arnaoutov, PhD, Staff Scientist
- Vasilisa Aksenova, PhD, Visiting Fellow
- Ming-Ta Lee, PhD, Visiting Fellow
- Sarine Markossian, PhD, Visiting Fellow
- Min Mo, PhD, Visiting Fellow
- Shaofei Zhang, PhD, Visiting Fellow
- Maia Ouspenskaia, DVM, Biologist
- Joseph Bareille, BA, Predoctoral Visiting Fellow
- Jasper Thompson, BA, Postbaccalaureate Intramural Research Training Award Fellow
We are interested in mechanisms of chromosome segregation, defects in which lead to aneuploidy, i.e., an abnormal number of chromosomes. Several common birth defects, such as Down's syndrome, result from aneuploidy arising during meiotic cell divisions. Moreover, aneuploidy arising from mitotic divisions is a hallmark of many types of solid tumors. During interphase, chromosomes are enclosed within nuclei, and exchange of all molecules between this compartment and the rest of the cell occurs through nuclear pore complexes (NPCs). Surprisingly, NPC proteins and proteins involved in trafficking of molecules into and out of the nucleus have important roles in chromosome segregation; we are investigating these roles at a molecular level. Our studies concentrated on a GTPase called Ran and on a family of small ubiquitin-like modifiers (SUMOs), which are indispensable for mitotic chromosome segregation. We also recently reported that the IRBIT protein is an inhibitor of ribonucleotide reductase; IRBIT works through a novel mechanism and is vital for genomic integrity.
The ultimate goals of our studies are to understand how these pathways enable accurate chromosome segregation and to discover how they are coordinated with each other and with other aspects of cell physiology.
Mitotic roles of nuclear pore complex proteins
NPCs consist of about thirty distinct proteins called nucleoporins. Kinetochores are proteinaceous structures that assemble at the centromere of each sister chromatid during mitosis and serve as sites of spindle microtubule attachment. The relationship between NPCs and mitotic kinetochores is surprisingly intimate but poorly understood. During interphase, several kinetochore proteins stably bind to NPCs (e.g., Mad1, Mad2, Mps1). During mitosis, metazoan NPCs disassemble, and at least a third of nucleoporins associate with kinetochores, including the RanBP2 complex and the Nup107-160 complex. We showed that the complexes play important roles in kinetochore function. Additional nucleoporins that do not associate with kinetochores have also been shown to have important mitotic roles, including Nup214, Nup98, and TPR.
Much of our current work concerns the RanBP2 complex. The mammalian RanBP2 complex consists of RanBP2 (a large nucleoporin that is also known as Nup358), SUMO-1–conjugated RanGAP1 (the activating protein for the Ran GTPase), and Ubc9 (the conjugating enzyme for the SUMO family of ubiquitin-like modifiers). The mammalian RanBP2 complex associates with kinetochores in a microtubule-dependent manner that also requires Crm1, a Ran-dependent nuclear export receptor. Disruption of RanBP2 association with kinetochores causes defective mitotic spindle assembly. Additional observations suggest an in vivo role of RanBP2 in interphase microtubule organization. We are also analyzing the association of RanBP2 and RanGAP1 in invertebrate species, particularly the fly Drosophila melanogaster. Flies mediate the association of RanBP2 and RanGAP1 through a biochemically distinct set of interactions that do not require SUMOylation or Ubc9. Elucidation of the mechanism of this association will not only allow us to test the importance of RanBP2 complex formation in a non-vertebrate system, but also provide a convenient alternative mechanism for the formation of this complex, to help us understand the complex's importance in vertebrate cells.
Mitotic regulation of the Ran GTPase
Ran is a Ras-family GTPase that plays critical roles in many cellular processes including nucleo-cytoplasmic transport, nuclear envelope assembly, and mitotic spindle assembly. Ran alternates between GDP– and GTP–bound forms. In interphase cells, GTP–bound Ran (Ran-GTP) is the major form in nucleus while GDP–bound Ran (Ran-GDP) is the predominant form in cytoplasm. The asymmetrical distribution of Ran-GTP and Ran-GDP drives cargo transport between the nucleus and cytoplasm through karyopherins, a family of nuclear transport carrier proteins that bind to Ran-GTP. In mitosis, after nuclear envelope breakdown, Ran-GTP is concentrated in the region close to mitotic chromatin, while Ran-GDP is the major form distal to chromatin. The Ran-GTP gradient guides mitotic spindle assembly by releasing spindle assembly factors (SAFs) from karyopherins based on local Ran-GTP concentrations. The conversion of Ran-GDP to Ran-GTP in cells is catalyzed by a Ran–specific guanine exchange factor (RanGEF), called RCC1 (Regulator of chromosome condensation 1) in vertebrates. The capacity of RCC1 to bind to chromatin establishes the asymmetrical distribution of Ran-GTP in interphase as well as the chromatin-centered Ran-GTP gradient in mitosis. Interestingly, RCC1’s association with chromatin is not static during the cell cycle and is regulated in a particularly dramatic fashion during anaphase in vertebrate systems. The regulation has not been correlated with post-translational modifications of RCC1, and the underlying molecular mechanism has not been reported.
RanBP1 is a highly conserved Ran-GTP–binding protein that acts as co-activator of RanGAP1 and can form a heterotrimeric complex with Ran and RCC1 in vitro. We found that RCC1 not associated with chromosomes during mitosis is sequestered and inhibited in RCC1/Ran/RanBP1 heterotrimeric complexes and that the sequestration is crucial for normal mitotic spindle assembly. In addition, RanBP1 complex formation competes with chromatin binding to regulate the distribution of RCC1 between the chromatin-associated and soluble fractions. Moreover, we identified a cell cycle–dependent phosphorylation on RanBP1 that modulates RCC1/Ran/RanBP1 heterotrimeric complex assembly and releases RCC1 to bind to chromatin; the phosphorylation is directly responsible for controlling RCC1 dynamics during anaphase. Together, our findings demonstrate novel roles of RanBP1 in spindle assembly and RCC1 regulation in mitosis (Reference 3) (Figure 1).
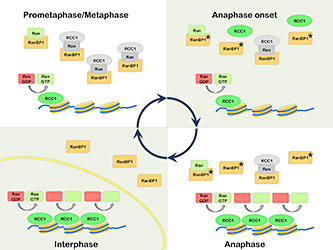
Click image to enlarge.
Figure 1. Model for mitotic regulation of RCC1 by RanBP1
Prometaphase: RCC1 is partitioned between an active, chromatin-bound pool (green) and an inactive pool (grey), associated with RRR complexes that also contain RanBP1 and nucleotide-free Ran (grey). Anaphase Onset: Phosphorylation of RanBP1 on Ser 60 (asterisk) releases RCC1 from the RRR complex. The free RCC1 is then recruited to chromatin. Anaphase: The high level of chromatin-bound RCC1 promotes enhanced levels of Ran-GDP (red) to Ran-GTP (green) exchange on chromatin. Interphase: Nuclear envelope (yellow) formation physically separates RCC1 from RanBP1, preventing RRR complex assembly and inhibition of RCC1.
SUMO–family small ubiquitin-like modifiers in higher eukaryotes
SUMOs are ubiquitin-like proteins (Ubls) that become conjugated to substrates through a pathway that is biochemically similar to ubiquitination. SUMOylation is involved in many cellular processes, including DNA metabolism, gene expression, and cell-cycle progression. Vertebrate cells express three major SUMO paralogs (SUMO-1–3): mature SUMO-2 and SUMO-3 are 95% identical to each other, while SUMO-1 is 45% identical to SUMO-2 or SUMO-3 (where they are functionally indistinguishable, we collectively call SUMO-2 and SUMO-3 SUMO-2/3). Like ubiquitin, SUMO-2/3 can be assembled into polymeric chains through the sequential conjugation of SUMOs to each other. Many SUMOylation substrates have been identified. SUMOylation promotes a variety of fates for individual targets, dependent upon the protein itself, the conjugated paralog, and whether the conjugated species contains a single SUMO or SUMO chains.
SUMOylation is dynamic owing to rapid turnover of conjugated species by SUMO proteases. Both post-translational processing of SUMO polypeptides and deSUMOylation are mediated by the same family of proteases that play a pivotal role in determining the spectrum of SUMOylated species. This group of proteases is called Ubl–specific proteases (Ulp) in yeast and Sentrin-specific proteases (SENP) in vertebrates. There are two yeast Ulps (Ulp1p and Ulp2p/Smt4p) and six mammalian SENPs (SENP1, SENP2, SENP3, SENP5, SENP6, and SENP7). SENP1, SENP2, SENP3, and SENP5 form a Ulp1p–related sub-family, while SENP6 and SENP7 are more closely related to Ulp2p. Yeast Ulps have important roles in mitotic progression and chromosome segregation. We defined the enzymatic specificity of the vertebrate SENP proteins and analyzed their key biological roles.
Ulp1p localizes to NPCs and is encoded by an essential gene; it is important for SUMO processing, nucleocytoplasmic trafficking, and late steps in the ribosome biogenesis pathway. Humans possess two NPC–associated SENPs: SENP1 and SENP2. While SENP2 is dispensable for cell division, mammalian SENP1 was recently shown to play an essential role in mitotic progression. Notably, we found that frogs possess a single NPC–bound SUMO protease, xSENP1, allowing us to examine the function of SUMO proteases at the NPC through analysis of a single protease (Reference 4). We determined the interaction partners of the enzyme throughout the cell cycle, using Xenopus egg extracts (XEEs). We found that xSENP1 associates strongly with Psmd1, the largest subunit of the proteasome 19S–regulatory particle (19S-RP). Proteasomes are complex ATP–dependent proteases that mediate the degradation of many cellular proteins typically targeted for destruction by ubiquitination. Ubiquitinated degradation substrates are fed into the proteasome’s catalytic 20S core particle (20S-CP) through the 19S-RP. Psmd1 plays a key structural role in the 19S-RP and acts as a binding site for the recruitment of other proteasome subunits, including Adrm1. Adrm1 is one of two subunits that directly recruit ubiquitinated substrates to the proteasome. While many proteasomal subunits have been found in proteomic screens for SUMOylation substrates, no role of these modifications has been reported. We mapped SUMOylation sites within Psmd1 and found that modification of a critical lysine adjacent to the Adrm1–binding domain regulates Adrm1 association with Psmd1. Our findings suggest that Psmd1 SUMOylation controls proteasome composition and function, providing a new mechanism for the regulation of ubiquitin-mediated protein degradation through the SUMO pathway. We speculate that the mechanism may allow the regulated degradation of critical mitotic substrates (Figure 2) and are currently testing the model.
Yeast Ulp2p is nucleoplasmic and not essential for vegetative growth but important for chromosome segregation. Ulp2p acts particularly in disassembly of poly-SUMO chains. We demonstrated that human SENP6 is a vertebrate Ulp2p–related enzyme that similarly prefers substrates containing multiple SUMO-2/3 moieties. We analyzed the mitotic role of SENP6 and found it to be essential for accurate chromosome segregation. Mitotic defects observed in the absence of SENP6 reflected the loss of inner kinetochore proteins, including components of the CENP-H/I/K/M and CENP-O centromere protein complexes. The findings demonstrate a novel function of the SUMO pathway in inner kinetochore assembly, which finely balances the incorporation and degradation of components of the kinetochore's inner plate. We are currently analyzing, at a biochemical level, how SUMO controls the deposition of CENP-H/I/K/M.
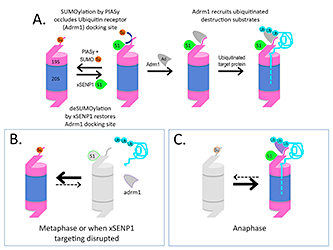
Click image to enlarge.
Figure 2. Working model for how SUMOylation regulates Adrm1 binding to the proteasome
A. Psmd1 becomes SUMO2/3 (Su)–conjugated by the SUMO ligase PIASy on Psmd1's C-terminus (shown as an extension from 19S-RP), causing occlusion of the Adrm1 (Ad)–binding site. The modification is antagonized by active xSENP1 (S1). Removal of the SUMO allows Adrm1 recruitment and the degradation of Adrm1–dependent ubiquitinated substrates (Ub). B. When xSENP1 targeting is disrupted, Adrm1 may be unable associate with the 19S-RP, depriving the proteasome of the ability to degrade a subset of targets. We are currently testing whether this mechanism controls proteasome activity during metaphase. C. We will also test whether changes upon anaphase onset could promote the degradation of important mitotic substrates through enhanced Psmd1 deSUMOylation.
IRBIT, a ribonucleotide reductase inhibitor with essential roles is genomic stability
Ribonucleotide reductase (RNR) provides deoxynucleotide triphosphates (dNTPs) for genomic and mitochondrial DNA replication and repair. Uncontrolled RNR activity has been associated with malignant transformation and tumor cell growth. RNR is subject to allosteric regulatory mechanisms in all eukaryotes, as well as to control by small protein inhibitors in budding and fission yeast. The key role of RNR in DNA synthesis has made it a target for both anticancer and antiviral therapy.
We discovered that the human IRBIT protein forms a dATP–dependent complex with RNR (Reference 5). The interaction appears to be closely conserved across a wide variety of metazoan species. dATP binds to the large subunit of RNR at two sites: dATP binding at the low-affinity activity site (A-site) inhibits RNR, while dATP binding to the specificity site (S-site) alters the enzyme’s substrate preferences. We found that A-site occupancy by dATP is critical for IRBIT–RNR binding, suggesting that IRBIT recognizes inactive, dATP–bound RNR subunit R1 and stabilizes the enzyme in its inhibited state. Notably, formation of the IRBIT–RNR complex is highly dependent upon the phosphorylation status of IRBIT under physiological conditions.
HeLa cells showed an imbalanced pool of dNTP after IRBIT depletion (Figure 3A). We found that this effect was most pronounced during the mitotic phase of the cell cycle, but that dNTP pools were less sensitive during G1 phase. Consistent with these findings, we observed that IRBIT bound to RNR more strongly during mitosis than during G1 phase. Live imaging of tissue culture cells demonstrated that IRBIT depletion causes an acceleration of the cell cycle, as well as much greater variation in between individual cells in overall cell cycle duration (Figure 3B). As observed in previous reports testing the consequences of dNTP pool perturbation, IRBIT loss was associated with changes in DNA replication patterns and disruption of genomic stability.
Together, our findings demonstrate an IRBIT–dependent mechanism for RNR regulation in higher eukaryotes, which acts by enhancing allosteric RNR inhibition by dATP (Figure 3C). We are currently working to understand the physiological role of IRBIT in the context of development and disease mechanisms and to exploit this mechanism to target RNR in therapeutic contexts.

Click image to enlarge.
Figure 3. IRBIT, a novel regulator of ribonucleotide reductase in higher eukaryotes
A. dNTP levels in control and IRBIT–depleted HeLa cells in mitotic (M) or G1 phases; a/s – asynchronous population. B. Cell-cycle progression of HeLa cells with or without IRBIT. The Y-axis indicates timing between anaphase and prometaphase of the next cell cycle of an individual daughter cell (n=20). Values are presented as the mean ± SEM. Statistical differences were evaluated by Student’s t test (P < 0.01). IRBIT-S68A is compromised in RNR binding because it cannot be phosphorylated within a critical regulatory motif. C. Schematic model for IRBIT inhibition of RNR. Top: RNR generates dNTPs, building blocks of DNA, from corresponding nucleotide precursors. Middle: dATP binds loosely to RNR and inactivates it. Bottom: IRBIT recognizes the RNR-dATP complex and stabilizes it by preventing dATP dissociation.
Phosphorylation of Xenopus p31comet by IKK-beta potentiates mitotic checkpoint exit.
Given that it prevents chromosome mis-segregation and averts genomic instability, the spindle assembly checkpoint (SAC) is among the most important cellular surveillance pathways. To do this, the SAC inhibits premature separation of mitotic sister chromatids by monitoring interactions between kinetochores (KTs) and spindle microtubules (MTs). SAC components are essential in vertebrate cells because organisms that undergo open mitosis must re-establish KT-MT attachments after nuclear envelope breakdown (NEBD) in each cell division. During the process, unattached KTs transiently invoke the SAC during the interval between NEBD and MT attachment, so that subsequent SAC silencing plays a key role in determining the timing of anaphase onset. While SAC silencing is thus critical for mitotic progression in metazoan cells, it remains poorly understood. The p31comet protein, found in higher eukaryotes, plays an important role in SAC silencing. We used Xenopus egg extracts (XEEs) to investigate the mitotic roles and regulation of p31comet. Our observations suggest that endogenous p31comet is important for anaphase timing in this system and is regulated through mitotic phosphorylation. While several well established mitotic kinases did not efficiently modify p31comet in vitro, IKK-beta (Inhibitor of nuclear factor k-B kinase-beta) was an effective p31comet kinase. Depletion or inhibition of IKK-beta delayed mitotic exit of XEEs, and a phosphomimetic p31comet mutant showed increased activity in SAC silencing. Together, our experiments suggest that p31comet contributes to the timing of anaphase onset in XEE through antagonism of the SAC and that IKK-beta modifies p31comet to enhance its activity. Several previous reports established that IKK-beta plays a clear but under-appreciated role of within mitosis, a role that is distinct from its function in cellular stress and inflammatory signaling pathways. Our findings are among the first mechanistic insights into the nature of this role.
Publications
- Mo M, Arnaoutov A, Dasso M. Phosphorylation of Xenopus p31comet potentiates mitotic checkpoint exit. Cell Cycle 2015; E-pub ahead of print.
- Ryu H, Yoshida MM, Sridharan V, Kumagai A, Dunphy WG, Dasso M, Azuma Y. SUMOylation of the C-terminal domain of DNA topoisomerase IIa regulates the centromeric localization of Claspin. Cell Cycle 2015; 14:2777-2784.
- Zhang MS, Arnaoutov A, Dasso M. RanBP1 governs spindle assembly by defining mitotic Ran-GTP production. Dev Cell 2014; 31:393-404.
- Ryu H, Gygi SP, Azuma Y, Arnaoutov A, Dasso M. SUMOylation of Psmd1 controls Adrm1 interaction with the proteasome. Cell Rep 2014; 7:1842-1848.
- Arnaoutov A, Dasso M. Enzyme regulation. IRBIT is a novel regulator of ribonucleotide reductase in higher eukaryotes. Science 2014; 345:1512-1515.
Collaborators
- Yoshiaki Azuma, PhD, University of Kansas, Lawrence, KS
- Chris Dealwis, PhD, Case Western Reserve University, Cleveland, OH
- Steven Gygi, PhD, Harvard Medical School, Boston, MA
- Vladimir Larionov, PhD, Developmental Therapeutics Branch, Center for Cancer Research, NCI, Bethesda, MD
- Andrea Musacchio, PhD, Max-Planck-Institut für Molekulare Physiologie, Dortmund, Germany
Contact
For more information, email mdasso@helix.nih.gov or visit http://sccr.nichd.nih.gov.


