Neuronal Circuits Controlling Behavior: Genetic Analysis in Zebrafish

- Harold Burgess, PhD, Head, Section on Behavioral Neurogenetics
- Sadie A. Bergeron, PhD, Postdoctoral Fellow
- Eric J. Horstick, PhD, Postdoctoral Fellow
- Kathryn M. Tabor, PhD, Postdoctoral Fellow
- Yared Bayleyen, BSc, Postbaccalaureate Fellow
- Mary R. Brown, BSc, Postbaccalaureate Fellow
- Gregory Marquart, BSc, Graduate student
- Jennifer L. Strykowski, MSc, Zebrafish Technician
Our goal is to understand how the nervous system selects appropriate motor responses, under diverse environmental contexts, in a way that best satisfies internal motivational objectives. We use zebrafish as a model because the larval brain exhibits the basic architecture of the vertebrate brain but is much less complex than the mammalian brain. Despite the relative simplicity of the nervous system, larvae have a sophisticated repertoire of sensory-guided and internally driven behaviors. In addition, the optical clarity of the embryo facilitates visualization of individual neurons and their manipulation with genetic techniques. Behavior in larvae is innate and therefore exhibits minimal variability between fish. Subtle alterations in behavior can therefore be robustly scored, making it possible to assess quickly the contribution of identified neurons to a variety of motor behaviors.
We focus on two aspects of behavioral regulation: the mechanisms by which sensory context regulates behavioral decisions; and pathways that sustain changes in behavioral state. In addition, we are developing a suite of genetic tools and behavioral assays to probe the nexus between neuronal function and behavior at single-cell resolution. The neuronal connections that allow the brain to integrate sensory and internal state information are established through genetic interactions during development. We aim to identify genes and neurons that are required for functional development of such connections. In vertebrates, neuronal circuits situated in the brainstem form the core of the locomotor control network and are responsible for balance, posture, motor control, and arousal. Accordingly, many neurological disorders stem from abnormal formation or function of brainstem circuits. Insights into the function of brainstem circuits in health and disease have come from genetic manipulation of neurons in zebrafish larvae in combination with computational analysis of behavior.
Molecular identification of neurons that mediate prepulse inhibition
Startle responses are rapid reflexes that are triggered by sudden sensory stimuli and help animals to defend against and escape from potential threatening stimuli. In both fish and mammals, startle responses are initiated by giant reticulospinal neurons in the medulla, which receive short-latency sensory input from diverse sensory modalities. Although highly stereotyped, startle responses are nevertheless modulated by sensory context and behavioral state and are therefore an excellent system in which to understand how such information is integrated with behavioral selection.
In characterizing how larvae respond to sensory cues, we observed well coordinated escape responses in response to a brief electric field pulse (EFP). The reaction time for EFP responses was remarkably short, leading us to question whether they were initiated by peripheral sensory neurons, as we had suspected. To address this question, we first determined whether the central neurons that drive escape responses are active during EFPs. We expressed the genetically encoded fluorescent calcium reporter GCaMP in reticulospinal neurons in order to monitor firing. As previously described, vibrational stimuli activated a broad network of reticulospinal neurons, including the Mauthner cell, the largest reticulospinal neuron in the larval brain. EFP stimuli also activated the Mauthner cell but failed to drive activity in other reticulospinal neurons. Ablation of the Mauthner cell completely abolished EFP responses, indicating that it is required for this behavior. Then, using genetic and pharmacological techniques, we isolated the Mauthner cell from synaptic input from other neurons, and found that it remained susceptible to activation by EFPs. We therefore proposed that EFPs bypass sensory cells and directly activate the Mauthner cell (Reference 1). This is likely because the Mauthner cell is a command neuron, which can drive an integrated behavioral response and has the largest diameter axon in the nervous system of the fish. Consistent with this, EFPs selectively triggered responses when larvae were aligned such that the longitudinal axis of the Mauthner axon was parallel to the field. Understanding the mechanism by which EFPs trigger escape responses now enables us to probe Mauthner cell excitability non-invasively so that we can measure how different sensory stimuli and behavioral states affect escape responsiveness (Reference 1).
In mammals, including humans, the startle response to a strong auditory stimulus can be inhibited by pre-exposure to a weak acoustic 'prepulse.' This form of startle modulation, termed prepulse inhibition, is diminished in several neurological conditions, including schizophrenia. The precise pattern of neuronal connections that enable prepulse inhibition is not known, yet understanding the mechanism of prepulse inhibition would permit rational investigation of how schizophrenia risk factors influence neuronal circuitry. Vibrational stimuli trigger rapid-escape swims in zebrafish that are mediated by giant reticulospinal neurons similar to the central neurons controlling startle responses in mammals. Escape swims are suppressed by pre-exposure to a prepulse, allowing us to apply the powerful suite of genetic tools in zebrafish to identify neurons that mediate prepulse inhibition.
To identify a transgenic zebrafish line that genetically labels neurons required for prepulse inhibition, we conducted a circuit-breaking screen. In this screen, we first generated a library of neuron-specific Gal4–enhancer trap lines marking distinct populations of neurons in the brain, then ablated the neurons in each enhancer trap line before testing for prepulse inhibition. We identified a transgenic line, y252, that labeled a discrete population of neurons in the hindbrain whose ablation or optogenetic inhibition led to dysregulation of prepulse inhibition. Dorsally located in the hindbrain, y252 neurons sent ventral projections toward the lateral dendrite of the Mauthner cell, the command neuron for escape responses in fish. We analyzed expression of neurotransmitter markers in y252 neurons and found that the neurons are located within one of four longitudinal columns of glutamatergic neurons extending through the rostro-caudal axis of the hindbrain. Moreover, blockade of NMDA receptors with MK801 phenocopied the effect of y252 neuron ablation, confirming that glutamatergic signaling is a central part of the mechanism for prepulse inhibition. Next, we discovered that the neurons genetically labeled in y252 are specified by Gsx1, a transcription factor previously implicated in differentiation of both excitatory and inhibitory interneurons in the spinal cord. Gsx1 was expressed in the mouse brainstem in the proliferative zone that gives rise to regions previously implicated in prepulse inhibition. We obtained Gsx1 knockout mice and performed behavioral testing. Knockout mice showed normal startle sensitivity but a strong reduction in prepulse inhibition. The results thus show that Gsx1 expression defines neurons that are required for prepulse inhibition across vertebrate species. Given that prepulse inhibition is abnormal in neuropsychiatric disorders with developmental origins, including schizophrenia and autism, our work will help identify and probe fundamental defects in circuitry abnormal in these conditions (Reference 2). We have now produced gsx1–mutant zebrafish, allowing us to analyze how defects in molecular signaling affect the development of prepulse inhibition circuitry.
Tools for analyzing neuronal circuits that control behavior
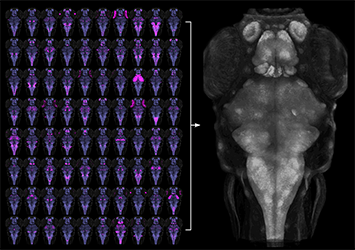
Click image to enlarge.
Figure 1. Registration of transgenic lines to a common reference brain
To aid functional neuroanatomical analysis in zebrafish, we scanned 80 Gal4 transgenic lines (left, magenta) at single micron resolution, to characterize cellular level expression patterns. We then computationally registered brain scans to a common reference brain (right).
We previously described a new method for restricting transgene expression to the nervous system, by incorporating a neuronal-restrictive silencing element. Using this method, we constructed a library of 240 Gal4 enhancer-trap transgenic fish with unique and restricted patterns of neuronal expression. We have now performed high-resolution brain imaging on more than 300 larvae, including 80 of these lines and 29 other transgenic lines. Using software developed for human brain imaging, we aligned the brain scans to a common reference, so that the expression in any of these lines can be compared at cellular level resolution (Figure 1). We then wrote brain browser software, which allows users to navigate within the larval zebrafish brain and to identify a transgenic line that labels any selected region (Figure 2). This is a powerful resource for neurobiological studies, as the 109 lines included in the database together provide a way to transgenically target more than 70% of the larval brain. Additionally, the software enables users to identify multiple transgenic lines that target the same cells, which will be useful for future studies using intersectional transgenic methods for targeting small populations of neurons (Reference 3).
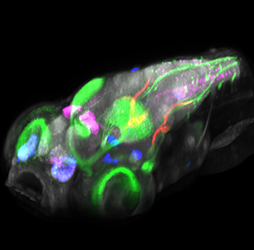
Click image to enlarge.
Figure 2. Brain browser software for visualizing expression of transgenes in larval zebrafish
3D projection view of a larval zebrafish, showing the expression of five transgenes: pan-neuronal Cerulean (grey), islet2b:GFP (green), and enhancer trap lines that label small populations of neurons (magenta, blue, and red). The lab imaged more than 100 transgenic lines at high resolution and computationally registered images to a common reference brain, so that expression patterns could be overlaid. The software allows transgenic lines that label selected brain regions to be quickly identified and permits prediction of transgenic lines that co-express in the same cell populations.
To complement these Gal4 lines that genetically target reporter genes to specific groups of neurons, we also built a set of UAS (upstream activation sequence) reporter lines. To optimize expression from our UAS lines, we analyzed DNA sequence features of genes that are highly expressed in zebrafish and developed an algorithm for optimizing transgene expression in zebrafish. We used the method to build strongly expressing UAS reporter lines to monitor and manipulate neurons and we also generated transgenic UAS lines for two novel genetic tools. First, through codon optimization and introduction of amino acid substitutions, we developed a significantly more active version of the bacterial nitroreductase gene (epNTR) that is used for cellular ablation (Reference 4). To complement this loss-of-function approach, we also tested overexpression of the voltage-gated sodium channel SCN5 in order to sensitize neurons to synaptic input. After SCN5 expression in motor neurons, larvae showed larger angle escape responses but normal startle sensitivity, and conversely, after SCN5 expression in the Mauthner cell, larvae showed normal escape kinematics but a reduced response threshold (References 1 and 5). Together, the methods enable us to interrogate neuronal cell function in vivo and assess their contribution to behavior. In addition, as many useful Gal4 lines show expression outside the nervous system, we developed a synthetic 3′ untranslated region incorporating microRNA binding sites that suppress reporter expression in heart, muscle, and skin (Figure 3) (Reference 3).
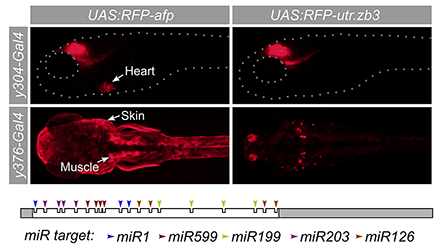
Click image to enlarge.
Figure 3. A synthetic 3′ untranslated region that suppresses expression outside the brain
Many Gal4 transgenic lines show useful expression in brain regions coupled with non-neuronal expression, such as y304 (heart) and y376 (muscle and skin). To suppress expression outside the brain, we designed a novel 3′ untranslated region (utr.zb3) that contains microRNA target sites for microRNAs expression in muscle, skin, and brain. A UAS (upstream activating sequence) reporter that expresses TagRFPT coupled to the new 3′ untranslated region strongly reduces expression in these tissues (right panels).
Additional Funding
- US Israel Binational Fund 2013433 (2014): Ongoing, with Dr Yoav Gothilf, Tel Aviv University
Publications
- Tabor K, Bergeron SA, Horstick EJ, Jordan DC, Aho V, Porkka-Heiskanen T, Haspel G, Burgess HA. Direct activation of the Mauthner cell by electric field pulses drives ultrarapid escape responses. J Neurophysiol 2014; 112:834-844.
- Bergeron SA, Carrier N, Li GH, Ahn S, Burgess HA. Gsx1 expression defines neurons required for prepulse inhibition. Mol Psychiatry 2015; 20:974-985.
- Marquart GD, Tabor KM, Brown M, Strykowski JL, Varshney GK, LaFave MC, Mueller T, Burgess SM, Higashijima I, Burgess HA. A 3D searchable database of transgenic zebrafish Gal4 and Cre lines for functional neuroanatomy studies. Front Neural Circuits 2015; 9:78.
- Horstick EJ, Jordan DC, Bergeron SA, Tabor KM, Serpe M, Feldman B, Burgess HA. Increased functional protein expression using nucleotide sequence features enriched in highly expressed genes in zebrafish. Nucleic Acids Res 2015; 43:e48.
- Horstick EJ, Tabor KM, Jordan DC, Burgess HA. Genetic silencing and selective activation using nitroreductase and TTX insensitive channels. Methods Mol Biol: Zebrafish 2015; in press.
Collaborators
- Kevin Briggman, PhD, Circuit Dynamics and Connectivity Unit, NINDS, Bethesda, MD
- Benjamin Feldman, PhD, Zebrafish Core Facility, NICHD, Bethesda, MD
- Yoav Gothilf, PhD, Tel-Aviv University, Tel-Aviv, Israel
- Shin-ichi Higashijima, PhD, National Institutes of Natural Sciences, Aichi, Japan
- Michael J. Iadarola, PhD, Warren Grant Magnuson Clinical Center, NIH, Bethesda, MD
- Thomas Mueller, PhD, Kansas State University, Manhattan, KS
- Ralph Nelson, PhD, Basic Neurosciences Program, NINDS, Rockville, MD
- Forbes Porter, MD, PhD, Program in Developmental Endocrinology and Genetics, NICHD, Bethesda, MD
- Mihaela Serpe, PhD, Program in Cellular Regulation and Metabolism, NICHD, Bethesda, MD
Contact
For more information, email haroldburgess@mail.nih.gov or visit http://ubn.nichd.nih.gov.


