Structural and Chemical Biology of Membrane Proteins

- Anirban Banerjee, PhD, Head, Unit on Structural and Chemical Biology of Membrane Proteins
- Eric Christenson, PhD, Postdoctoral Fellow
- Raffaello Verardi, PhD, Postdoctoral Fellow
- Jin-Sik Kim, PhD, Visiting Fellow
- Pramod Kumar, PhD, Visiting Fellow
- Chul-jin Lee, PhD, Visiting Fellow
- Mitra Rana, PhD, Visiting Fellow
Cell membranes lie at the heart of cellular compartmentalization. Integral membrane proteins, which are embedded in cell membranes, perform critically important functions, exemplified by the propagation of electrical signals along the cellular surface, exchange of material between two cellular compartments, and response of a cell to numerous signaling cues. The biomedical relevance of membrane proteins is underscored by the fact that more than 50% of drug targets are membrane proteins. We are interested in the structural basis of the function of several integral membrane protein families. We combine high-resolution structural techniques such as X-ray crystallography with functional analyses using a range of biochemical and biophysical techniques. In addition to solving high-resolution structures and using them to guide functional experiments, we are also working on discovering novel small molecule probes for our target proteins using a combination of computational and experimental approaches. Not only are our target proteins critically important to a range of physiological processes, they have been linked to several human diseases, most notably neuropsychiatric diseases, various forms of cancer, and protoporphyria. Through our biochemical and biophysical studies, we are providing the first ever detailed molecular mechanisms of their functions that, in turn, are leading to novel hypotheses that can be tested experimentally. Through our small-molecule discovery program, we hope to develop new tools to investigate the cellular functions of our target proteins as well as to permit the development of novel therapies for human diseases.
Molecular mechanism of post-translational protein lipidation by DHHC palmitoyltransferases
Covalent modification of proteins by attachment of fatty acids to internal cysteines through acyl linkages constitutes one of the most pervasive and physiologically important post-translational modifications. These are referred to collectively as protein palmitoylation, or more generally, protein S-acylation. The repertoire of palmitoylated proteins has expanded rapidly in recent years, with hundreds of new members being added to the cellular “palmitoylome.” The physico-chemical effect of palmitoylation is to alter the local hydrophobicity of the substrate protein. The thioester bond makes S-acylation unique in that it is a labile moiety and can be cleaved, in the cellular context, by thioesterase enzymes (Figure 1). Thus S-acylation is one of the few dynamic post-translational modifications. The physiological effects of S-acylation are diverse and have critical cellular importance; Ras, a small GTPase that is critical for cellular growth and differentiation and mutated in about one-third of all human cancers, is palmitoylated at the Golgi and is subsequently targeted to the plasma membrane by vesicular transport. Palmitoylated Ras localizes to cholesterol-rich domains on the plasma membrane. However, it subsequently gets depalmitoylated by the thioesterase APT1 and dissociates from the plasma membrane and redistributes on endomembranes including the Golgi. This dynamic recycling of Ras is critical for its function.
Protein S-acylation is catalyzed by a large group of enzymes known as DHHC-palmitoylacyltransferase (DHHC-PAT), so named because they contain a signature D-H-H-C motif in a cysteine-rich domain in an intracellular loop (Figure 1). These are low-abundance polytopic integral membrane proteins localized at a variety of cellular compartments. Yeast has seven DHHC-PATs and mammals have up to 24 putative DHHC-PATs encoded in their genome. Beyond the shared DHHC domain, DHHC-PATs vary considerably; some possess ankyrin repeats, a few have six transmembrane helices instead of four and at least one forms a functional heterodimer with a cytoplasmic auxiliary subunit (Figure. 1). To date, there are no reported consensus sequences for palmitoylation. A specific DHHC-PAT can palmitoylate multiple substrates, and conversely a given substrate can be palmitoylated by multiple DHHC-PATs. This redundancy has been one of the most intriguing aspects of DHHC-PATs and makes it difficult to assign substrates by overexpression/knockout strategies, given that in the absence of one enzyme, others can take over. However, this does not necessarily reflect the true enzyme-substrate relationship. The situation has been even more confounded by the lack of specific inhibitors of DHHC-PATs. Even though 2-bromopalmitate is widely used as a global inhibitor of DHHC-PATs, it has been shown that it broadly targets other proteins involved in lipid metabolism.
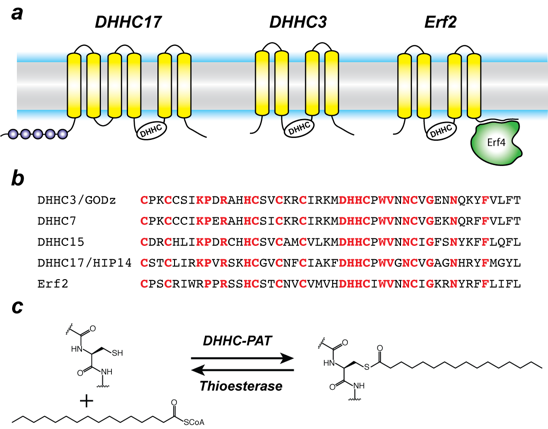
Click image to enlarge.
Figure 1. Organization and properties of DHHC palmitoyltransferases
a) The organization of three different DHHC-PATs are shown schematically. The spheres indicate protein-protein interaction domains. Erf2 associates with a cytoplasmic subunit, Erf4, to form the active enzyme.
b) The DHHC-CRD region of a few representative DHHC-PAT’s are aligned. The conserved amino acids are shown in red.
c) Reaction catalyzed by DHHC-PATs; the reverse reaction is catalyzed by acylprotein thioesterases (APT).
Besides its broad importance in cell biology, palmitoylation has been linked to several diseases, most notably neuropsychiatric disorders such as Huntingtin’s disease and various forms of cancer. Very recently, it was shown that DHHC20 palmitoylates epidermal growth factor (EGFR) and is thus a potential therapeutic target for a broad range of cancers. However, despite their importance across a broad spectrum of biological pathways and their biomedical importance, very little is known about the molecular mechanism of DHHC palmitoyltransferases. There is nothing known about their structural organization or how they interact with substrates and the fatty acyl CoA that serves as the acyl donor.
In the first breakthrough in this field, we solved a high-resolution crystal structure of the complex between the substrate recognition domain of human DHHC17 and a fragment of Snap25b, one of the canonical substrates of DHHC17. Through structure-guided mutagenesis, we discovered key residues in DHHC17 that are critically important for interaction with Snap25b. We further extended our finding by showing that the same residues are also crucial for the interaction of DHHC17 with Huntingtin, one of its most relevant substrates. We are currently studying other members of this intriguing family of enzymes to obtain a detailed molecular understanding of how they function.
Molecular mechanism of iron transport into mitochondria
The importance of iron in biology cannot quite be overstated. In higher organisms, mitochondria are the ‘hotspot’ for the cell biology of iron because this is where Fe-S clusters are biosynthesized and iron is inserted into heme. Mitochondrial iron homeostasis plays a critical role in cellular iron homeostasis and in the overall physiology of the cell. In vertebrates, the only known major transporters of iron into mitochondria are mitoferrin-1 and mitoferrin-2, two homologous members of a large group of mitochondrial transporters known as the Mitochondrial Carrier family (Figure 2). Mitoferrin-1 (Mfrn1) is expressed mainly in erythroid cells while mitoferrin-2 is expressed ubiquitously. Knockout of Mfrn1 is embryonically lethal, reflecting the importance of mitoferrins in vertebrate physiology.
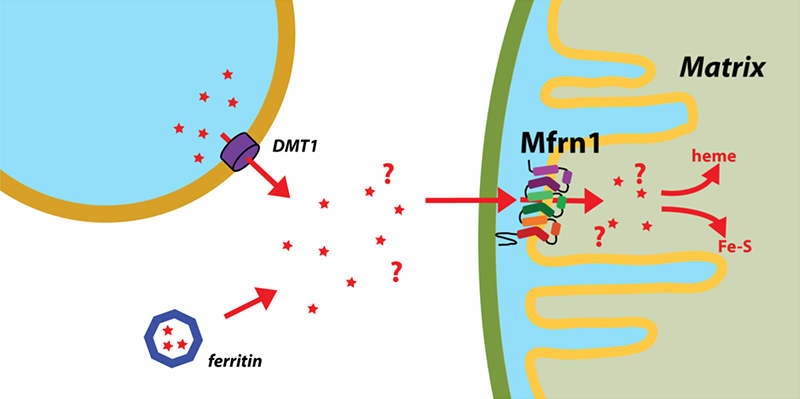
Click image to enlarge.
Figure 2. Mitoferrin and mitochondrial iron transport
Iron is transported out of endosomes into the cytoplasm by Divalent Metal Ion Transporter 1 and is subsequently directed to Mitoferrin by an unknown mechanism. Mitoferrins are the only known major transporters into mitochondria of the iron that is subsequently utilized for important processes such as heme and Fe-S cluster biosynthesis.
Mfrn1 and Mfrn2 were discovered more than 10 years ago. However, the proposed iron transport activity of either transporter has not yet been demonstrated using an in vitro functional reconstitution assay. Also no report about their interaction with iron or other related metal ions exists, most likely because heterologous overexpression and purification of mitoferrins have not yet been reported in literature. If mitoferrins do indeed transport iron, it is important to know how selective they are for iron over other similar metal ions, what residues in mitoferrin-1 and -2 are important for their function, and whether they even bind iron. These fundamental questions about mitoferrin function have, to date, not been addressed.
We have now carried out heterologous purification and in vitro functional reconstitution and mutational dissection of a vertebrate Mfrn1. This is the first demonstration that Mfrn1 can indeed transport iron. We showed that Mfrn1 is a promiscuous metal ion transporter in that it also transports other first-row transition metal ions. Through mutagenesis, we discovered candidate residues that are important for metal ion transport by Mfrn1 and those that could be involved in forming metal-ion binding sites during transport.
Our studies provide the first biochemical insights into Mfrn function and form the starting point for future high-resolution structural studies of Mfrn function. Our transport assay and the purification strategy will lead to more detailed biochemical and biophysical experiments into the mechanistic basis of iron transport by Mfrn1 and Mfrn2. Our in vitro proteoliposome-reconstituted iron and copper transport assay is likely to be used for studying other iron and copper transporters and, considering the importance of iron and copper transport in biology, these assays will be important tools for biochemical dissection of the transporters of these metal ions.
Additional Funding
- NIH Intramural AIDS Targeted Antiviral Program (IATAP) award to Anirban Banerjee (2017)
Publications
- Verardi R, Kim JS, Ghirlando R, Banerjee A. Structural basis for substrate recognition by the ankyrin repeat domain of human DHHC17 palmitoyltransferase. Structure 2017;25:1337-1347.
Collaborators
- Rodolfo Ghirlando, PhD, Laboratory of Molecular Biology, NIDDK, Bethesda, MD
- James Inglese, PhD, Assay Development & Screening Technology Laboratory, NCATS, Bethesda, MD
- Kanagalaghatta R. Rajashankar, PhD, NE-CAT, Argonne National Laboratory and Cornell University, Lemont, IL
- Lei Shi, PhD, Computational Chemistry and Molecular Biophysics Unit, NIDA, Baltimore, MD
Contact
For more information, email anirban.banerjee@nih.gov or visit http://banerjee.nichd.nih.gov.


