Cell Cycle Regulation in Oogenesis

- Mary Lilly, PhD, Head, Section on Gamete Development
- Kuikwon Kim, MS, Technician
- Lucia Bettedi, PhD, Visiting Fellow
- Shu Yang, PhD, Visiting Fellow
- Yingbiao Zhang Zhang, PhD, Visiting Fellow
- Elena Ghanaim, BS, Postbaccalaureate Fellow
- Nicholas Johnson, BS, Postbaccalaureate Fellow
- Christa Ventresca, BS, Postbaccalaureate Fellow
The long-term goal of our laboratory is to understand how the cell-cycle events of meiosis are coordinated with the developmental events of gametogenesis. Chromosome mis-segregation during female meiosis is the leading cause of miscarriages and birth defects in humans. Recent evidence suggests that many meiotic errors occur downstream of defects in oocyte growth and/or the hormonal signaling pathways that drive differentiation of the oocyte. Thus, an understanding of how meiotic progression and gamete differentiation are coordinated during oogenesis is essential to studies in both reproductive biology and medicine. We use the genetically tractable model organism Drosophila melanogaster to examine how meiotic progression is instructed by the developmental and metabolic program of the egg.
In mammals, studies on the early stages of oogenesis face serious technical challenges in that entry into the meiotic cycle, meiotic recombination, and the initiation of the highly conserved prophase I arrest all occur during embryogenesis. By contrast, in Drosophila these critical events of early oogenesis all take place continuously within the adult female. Easy access to the early stages of oogenesis, coupled with available genetic and molecular genetic tools, makes Drosophila an excellent model for studies on meiotic progression and oocyte development.
To understand the regulatory inputs that control early meiotic progression, we are working to determine how the oocyte initiates and then maintains the meiotic cycle within the challenging environment of the ovarian cyst. Our studies focus on the following questions, which are relevant to the development of all animal oocytes: the nature of strategies the oocyte uses to protect itself against inappropriate DNA replication; how the oocyte inhibits mitotic activity before meiotic maturation and the full growth and development of the egg; and how cell-cycle and metabolic status within the ovarian cyst influence the differentiation of the oocyte. To answer these questions, we have undertaken studies to determine the basic cell-cycle and metabolic program of the developing ovarian cyst.
The GATOR complex: integrating developmental and metabolic signals in oogenesis
We are interested in how metabolism influences oocyte growth, development, and quality. Target of Rapamycin Complex 1 (TORC1) is a primary regulator of cell growth and metabolism, which responds to several upstream signals, including nutrient availability, energy status, and growth factors. In the past year, we defined the role of the GATOR complex in the regulation of metabolic homeostasis, autophagic flux, endomembrane dynamics, and oocyte development. Our findings define Drosophila as an excellent model for the genetic dissection of the GATOR complex and its role in preventing the developmental defects and pathologies associated with deregulated TORC1 activity. Additionally, they clearly establish a conserved role for the GATOR complex in the regulation of meiotic progression and gametogenesis.
The GATOR complex comprises two sub-complexes. The GATOR1 complex inhibits TORC1 activity in response to amino acid starvation by preventing the targeting of TORC1 to lysosomes, where the complex is activated by the GTP–binding protein Rheb. GATOR1 is a trimeric protein complex, consisting of the proteins Nprl2, Nprl3, and Iml1. Recent evidence from yeast and mammals indicates that the components of the GATOR1 complex function as GTPase–activating proteins (GAP) that inhibit TORC1 activity by inactivating the Rag GTPases, which are required for TORC1 lysosomal recruitment. Notably, Nprl2 and Iml1 are tumor suppressor genes while mutations in Iml1, known as DEPDC5 in mammals, are the leading cause of hereditary epilepsy.
The GATOR2 complex comprises five proteins: Seh1, Sec13, Mio, Wdr24, and Wdr59. Our work, as well as that of others, found that the GATOR2 complex activates TORC1 by opposing the TORC1–inhibitory activity of GATOR1. Intriguingly, computational analysis indicates that Mio and Seh1, as well as several other members of the GATOR2 complex, have structural features consistent with coatomer proteins and membrane-tethering complexes. In line with the structural similarity to proteins that influence membrane curvature, we showed that three components of the GATOR2 complex, Mio, Seh1, and Sea2, localize to the outer surface of lysosomes, the site of TORC1 regulation. However, how GATOR2 inhibits GATOR1 activity, thus allowing for the robust activation of TORC1, remains unknown. Additionally, the role of the GATOR1 and GATOR2 complexes in both the development and physiology of multicellular animals remains poorly defined. Over the last year, we used molecular, genetic, and cell-biological approaches to define the role of the GATOR complex in the regulation of Drosophila development and physiology.
A genetic screen for new regulators of the GATOR complex
Mutations in the GATOR2 components mio and seh1 cause the constitutive activation of the GATOR1 complex in the female germ line, resulting in permanent inhibition of TORC1 activity and a block to oocyte growth and development. We determined that germline RNAi depletions of any of the GATOR1 components in the mio and seh1 mutant backgrounds relieve this permanent TORC1 inhibition and rescue the mio and seh1 ovarian phenotypes. Over the last year, we used this epistatic relationship to conduct a high-throughput RNAi–based screen to identify upstream regulators and downstream effectors of the GATOR1 complex during oogenesis. First, we determined that expressing a short hairpin RNA against the seh1 transcript, using the nanos-Gal4 germline-specific driver, recapitulates the seh1 mutant ovarian phenotype. Moreover, co-depleting nprl2, nprl3, and iml1 dramatically rescues the seh1RNAi ovarian phenotype (Figure 1). In order to identify additional genes that, when co-depleted with seh1, rescue the seh1RNAi ovarian phenotypes, we used RNAi lines from the Transgeneic RNAi Project (TRiP) that have been optimized for germline expression. This co-depletion screen identified an array of new GATOR–interacting genes. Importantly, as anticipated, the screen identified several known regulators of TORC1, including Tsc1, Tsc2, and PTEN. Additionally, we identified many genes that function in translation, transcription, and the DNA–damage response that had not previously been implicated in metabolic regulation. These newly identified GATOR–interacting genes will provide a framework for our future studies on the regulation and function of the GATOR complex during oogenesis.
Click image to enlarge.
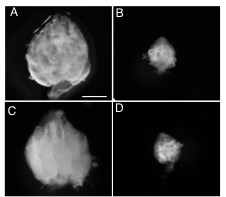
Figure 1. seh1 RNAi depletion and co-depletion phenotypes
Representative images of (A) wild-type (WT) (large ovary), (B) nanos>seh1RNAi (small ovary), (C) nanos>seh1RNAi, nprl2RNAi (large ovary), (D) nanos>seh1RNAi, mCherryRNAi (small ovary). Note that co-depletion of nprl2 rescues the seh1RNAi phenotype while co-depletion of mCherry fails to rescue the seh1RNAi phenotype.
The GATOR1 complex promotes genomic stability during the early meiotic cycle.
Meiosis must accomplish two seemingly incompatible goals. First, it must faithfully copy and distribute genetic material to the next generation. Second, it must promote genomic diversity through meiotic recombination, a process initiated by the production of DNA double-stranded breaks. In our previous work, we demonstrated that, in Drosophila females, the timely entry into meiosis requires the down-regulation of TORC1 by the GATOR1 complex. However, precisely why a nutrient sensor has a retained an essential role in the regulation of the early meiotic cycle in a metazoan remained unclear. One model posits that, in addition to promoting meiotic entry, the GATOR1 complex might facilitate other events of the early meiotic cycle such as the highly conserved events of meiotic recombination. The controlled generation of DNA double-stranded breaks by the Spo11 enzyme initiates meiotic recombination in early prophase of meiosis I.
We determined that the GATOR1 complex promotes genomic stability in the presence of meiotic double-stranded breaks in both Drosophila melanogaster and mouse. Specifically, GATOR1–mutant oocytes exhibit a dramatic increase in the steady-state number of double-stranded breaks (Figure 2). Moreover, in contrast to wild-type oocytes that repair their double-stranded breaks prior to oocyte growth, GATOR1–mutant oocytes retain double-stranded breaks into late states of oogenesis. In GATOR1–mutant ovarian cysts, the increase in unrepaired DNA double-stranded breaks is accompanied by an increase in p53 activity, as monitored by a p53 reporter construct. p53 is a highly-conserved transcription factor that mediates a response to genotoxic stress. Strikingly, inhibiting the generation of meiotic double-stranded breaks strongly suppressed p53 activity in the nprl3–/––mutant background. From these data, we conclude that the GATOR1 complex is required to maintain genomic stability in the presence of meiotic double-stranded breaks and is thus essential for oocytes to safely complete meiotic recombination. Finally, in line with a conserved function for the GATOR1 complex in mammals, we showed that the GATOR1 component DEPCD5/Iml1 regulates meiotic double-stranded breaks in mouse spermatocytes. Taken together, our data support a model in which the GATOR1 complex has retained a conserved role in the early meiotic cycle and early gametogenesis in metazoans.
Click image to enlarge.
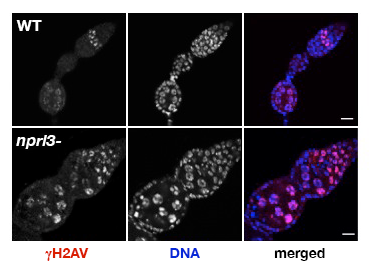
Figure 2. nprl3–mutant ovarian cysts have elevated levels of DNA damage.
Note that WT ovarian cysts rapidly repair meiotic double-stranded breaks, as indicated by the loss of gamma-H2Av in the early germarium. In contrast, nprl3 mutants retain high levels of gamma-H2Av into late stages of oogenesis.
The GATOR2 component Wdr24 has both TORC1–dependent and –independent functions.
While the GATOR1 complex has been implicated in a wide array of human pathologies, including cancer and hereditary forms of epilepsy, the in vivo relevance of the GATOR2 complex remains poorly understood in metazoans. We defined the in vivo role of the GATOR2 component Wdr24 in Drosophila. Using a combination of genetic, biochemical, and cell-biological techniques, we demonstrated that Wdr24 has both TORC1–dependent and TORC1–independent functions in the regulation of cellular metabolism. Through the characterization of a null allele, we found that Wdr24 is a critical effector of the GATOR2 complex required for the robust activation of TORC1 and cellular growth in a broad array of Drosophila tissues (Figure 3). Additionally, epistasis analysis between wdr24 and genes that encode components of the GATOR1 complex revealed that Wdr24 has a second critical function: the TORC1–independent regulation of lysosome dynamics and autophagic flux. Notably, we found that two additional members of the GATOR2 complex, Mio and Seh1, also play a TORC1–independent role in the regulation of lysosome function. The results represent a surprising and previously unrecognized function of GATOR2 complex components in the regulation of lysosome structure and function. Consistent with our findings in Drosophila, through the characterization of a wdr24–/– knockout HeLa cell line, we determined that Wdr24 promotes lysosome acidification and autophagic flux in mammalian cells. Taken together, our data support a model in which Wdr24 is a key effector of the GATOR2 complex, required for both TORC1 activation and the TORC1–independent regulation of lysosomes. Moreover, our data raise the interesting possibility that the GATOR2 complex regulates GATOR1 through the control of lysosome structure and/or function.
Click image to enlarge.
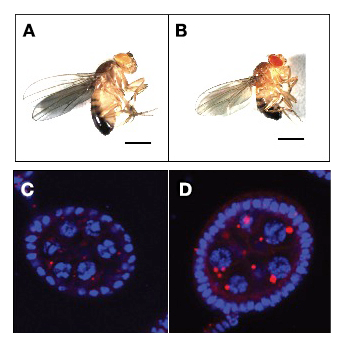
Figure 3. The GATOR2 component Wdr24 regulates growth and autophagy.
Representative images of (A) WT and (B) wdr241–mutant adult males. wdr241 males are notably smaller than WT males. Size bar is 100 μm. Representative images of (C) WT and (D) wdr241 egg chambers from well fed females stained with DAPI to mark DNA (blue) and LysoTracker (red) to mark autolysosomes. Note that wdr241 egg chambers accumulate large numbers of autolysosome-like structures under nutrient-replete conditions.
Publications
- Wei Y, Reveal B, Cai, W, Lilly MA. The GATOR1 complex regulates metabolic homeostasis and the response to nutrient stress in Drosophila. G3 (Bethesda) 2016 6(12):3859-3867.
- Cai W, Wei Y, Jarnik M, Reich J, Lilly MA. The GATOR2 component Wdr24 regulates TORC1 activity and lysosome function. PLoS Genetics 2016 12(5):e1006036.
Collaborators
- Juan Bonifacino, PhD, Section on Intracellular Protein Trafficking, NICHD, Bethesda, MD
- Brian Calvi, PhD, Indiana University, Bloomington, IN
- Daniel Camerini-Otero, PhD, Genetics and Biochemistry Branch, NIDDK, Bethesda, MD
Contact
For more information, email mlilly@helix.nih.gov or visit http://cbmp.nichd.nih.gov/uccr.


