Molecular Genetics of Embryogenesis in Zebrafish and Xenopus

- Igor B. Dawid, PhD, Head, Section on Developmental Biology
- Alison M. Heffer, PhD, Intramural Research Training Award Fellow
- Allisan Aquilina-Beck, MS, Fish Technician
The laboratory uses the frog Xenopus laevis and the zebrafish Danio rerio as experimental systems in which to study molecular-genetic mechanisms of early vertebrate development. Recently, we focused on the mechanisms and role of protein stability in regulating early pancreas development and on the function of the BTB domain–containing protein Kctd15 in development. The BTB domain is a protein-protein interaction domain that is part of many proteins. Understanding its function in various contexts will help elucidate the mechanisms of protein interactions.
The BTB–domain protein Kctd15 is an inhibitor of neural crest formation and of the transcription factor AP-2.
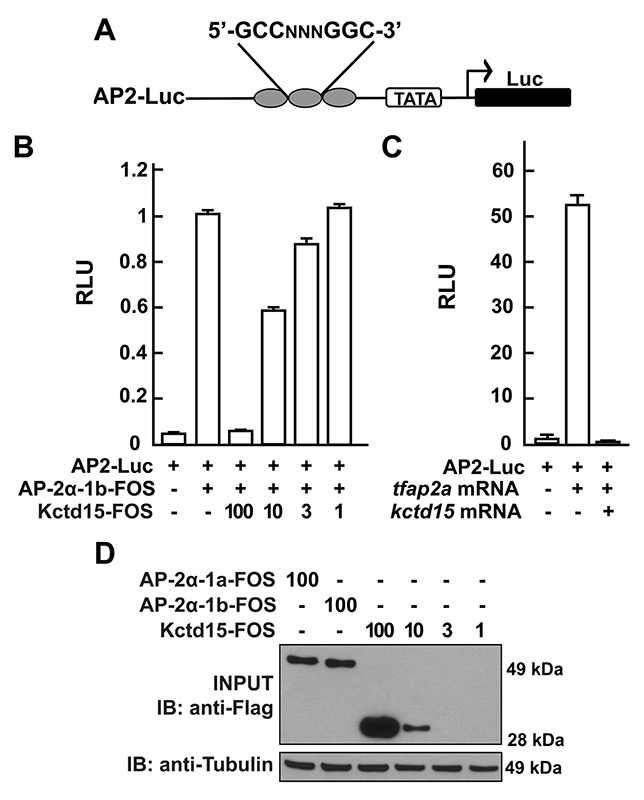
Click image to enlarge.
Figure 1. Kcdt15 represses AP-2 function.
(A) AP2-Luc reporter, containing three AP-2 consensus sites driving expression of luciferase (Luc).
(B) The reporter was strongly stimulated by zebrafish AP-2α and dramatically inhibited by zebrafish Kctd15 (levels indicated in ng). RLU, relative light units.
(C) Reporter activity in zebrafish embryos.
(D) Cells were transfected with FOS–tagged zebrafish AP-2α and Kctd15, and lysates were blotted to assay expression of both proteins.
From Reference 1.
Our laboratory has a long-standing interest in the formation of the neural crest (NC), a group of cells with stem-cell properties that arise at the dorsal neural tube and migrate to many locations in the embryo to give rise to a large number of varied differentiated derivatives. A recent focus was the role of the BTB domain–containing protein Kctd15, which is capable of inhibiting NC formation. In pursuing the molecular mechanism of Kctd15 action, we found that Kctd15 regulates the activity of the transcription factor AP-2. AP-2 is known to play a key role in the induction and differentiation of NC cells. We found that AP-2 and Kctd15 can interact when co-expressed in cultured cells. Further, Kctd15 is a highly effective inhibitor of AP-2 activity in a reporter assay (Figure 1). In studying the mechanism of inhibition of AP-2 by Kctd15, we found that Kctd15 interacts with the activation domain of AP-2. We further analyzed this system using a fusion product between the Gal4 DNA–binding domain and the AP-2–activation domain. Within the latter domain, a conserved proline-rich motif proved critical for Kctd15 interaction: mutation of proline 59 to alanine (P59A) in the Gal4–AP-2 fusion resulted in a protein that was active but could not bind and was insensitive to inhibition by Kctd15 (Figure 2). The proline residue was also essential for Kctd15 sensitivity in the context of the full-length AP-2 molecule. Thus, we conclude that Kctd15 inhibits AP-2 by binding to a specific site in its activation domain (Reference 1).
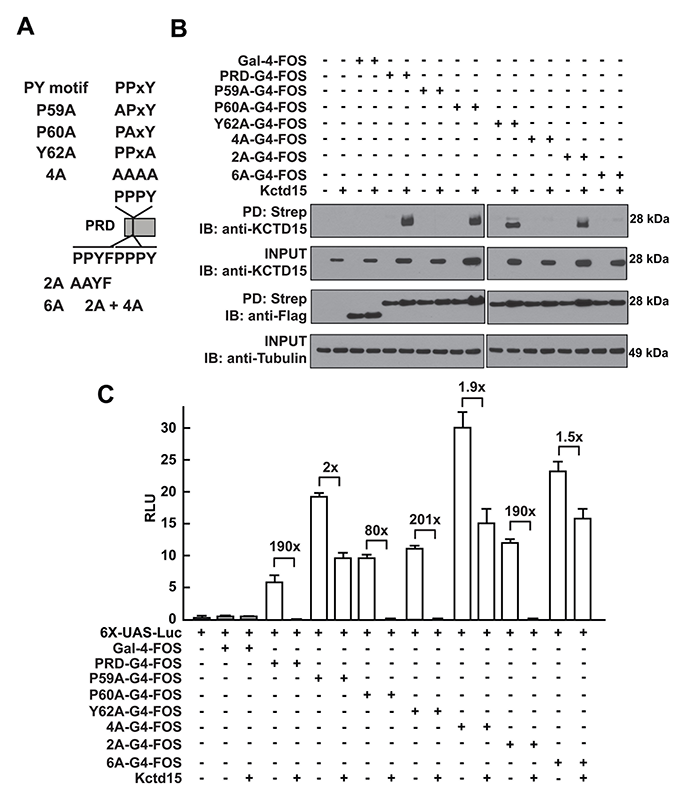
Click image to enlarge.
Figure 2. Critical role of P59 for Kctd15 interaction and inhibition of AP-2
(A) Mutants in the conserved PPxY motif within the AP-2α proline-rich domain (PRD) and two additional mutants in the adjoining region are shown. PRDs were fused to the Gal4-DNA–binding domain and the FOS tag. (B) Kctd15 interaction depends on proline 59 (P59). WT and mutant fusion constructs were expressed in HEK293T cells alone or with Kctd15, and complexes were pulled down using a Strep-Tactin matrix. PRDs could bind to Kctd15 except in the constructs containing the P59A mutation (P59A, 4A and 6A). The constructs were expressed at similar levels. (C) PPxY mutants activate the UAS-Luc reporter two-to-five fold more highly than does WT PRD-Gal4. PRD-Gal4 WT and P60A, Y62A (tyrosine 62 to alanine), and 2A were dramatically inhibited by zebrafish Kctd15, whereas P59A and mutants containing this change were inhibited two-fold or less. Figure from Reference 1.
Recently, we tested for Kctd15 function by overexpressing this protein in animal explants (animal caps) from Xenopus embryos, followed by microarray analysis of the transcriptome of control and overexpressing explants. We confirmed that Kctd15 inhibits several markers of the neural crest, and we reported on additional genes affected by overexpression of this factor (Reference 5).
In addition, we generated frameshift mutations in both kctd15 paralogs in zebrafish, using TALENs restriction enzymes for gene editing. Mutants in both kctd15a and kctd15b are viable and fertile but show slow growth and several additional phenotypes, which are currently under study.
Lnx2 ubiquitin ligase is essential for exocrine cell differentiation in the early zebrafish pancreas.
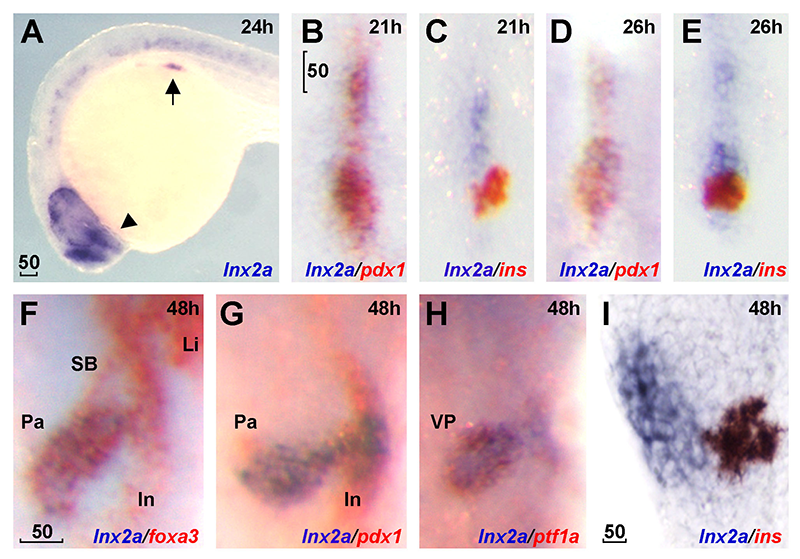
Click image to enlarge.
Figure 3. lnx2a is expressed in the ventral pancreas during early pancreas specification.
Lateral views (A and I), dorsal views (B–E), and ventral views (F–H). (A) lnx2a transcripts (blue) are detected in regions of the endoderm (arrow), forebrain (arrowhead) and the spinal cord at 24 hours post fertilization (hpf). Expression of pdx1 (B, D, and G), ins (C, E, and I) and foxa3 (F) are in red. lnx2a expression is observed in the antero-ventral part of pdx1–positive pancreas precursors at 21 (B) and 26 (D) hpf, but excluded from β-cells (ins+) in the dorsal pancreas at 21 (C), 26 (E), and 48 (I) hpf. At 48 hpf, lnx2a expression is detected in the ventral pancreas but not in the intestine, swim bladder, or liver (F–H). Li, liver; In, intestine; Pa, pancreas; SB, swim bladder; VP, ventral pancreas; scale bar, 50μm.
Pancreas development is of great interest as a prime example of organogenesis and because of the importance of understanding the development of insulin-producing cells and regulation of insulin synthesis. We found that the E3 ubiquitin ligases Lnx2a and Lnx2b play an important role in early pancreas differentiation in the zebrafish. While mammals have only one Lnx2 gene, zebrafish have two closely related paralogs, lnx2a and lnx2b. In studying the gene that encodes Lnx2a, we found that it is expressed in the ventral pancreatic bud, in addition to being expressed in the nervous system (Figure 3). In the early zebrafish embryo, the ventral bud gives rise to exocrine cells and the dorsal bud to the primary islet, which contains endocrine cells. In later development, ventral bud progenitors give rise to secondary islets and duct cells in addition to exocrine cells. We found that a splice morpholino (MO) that blocks lnx2a exon2/3 splicing inhibits exocrine marker expression while leaving endocrine marker expression unaffected (Figure 4). To test MO specificity, we used TALEN–mediated gene targeting to generate a frame-shift mutation, lnx2adelta70, that does not produce Lnx2a protein. The mutant fish proved to be phenotypically wild type (WT), with normal exocrine gene expression (Figure 5A–E). Further analysis showed that the splice MO led to exon skipping, generating an mRNA that produces a protein shorter than the WT (Figure 5F–H). Mass spectrometry showed the protein to be an N-truncated version of the WT protein that had lost the catalytically active RING domain. We wondered whether the truncated protein could act as an interfering factor and found this to be the case, as injection of RNA encoding the truncated protein into lnx2adelta70 embryos reproduced the exocrine deficiency phenotype (Figure 5I and J). The effect is the result of inhibition of Lnx2b function by the truncated Lnx2a, as indicated by the fact that injection of lnx2b-MO into lnx2adelta70 reproduced the exocrine deficiency phenotype (Figure 5D and E). Definitive genetic evidence for our interpretation of the mechanism of the lnx2a-splice MO phenotype came from a mutation, lnx2adelta329, that deletes the exon/intron boundary targeted by the splice MO. lnx2adelta329–mutant fish effectively reproduce the exocrine deficiency phenotype seen originally in lnx2a-splice MO–injected embryos (Figure 6).
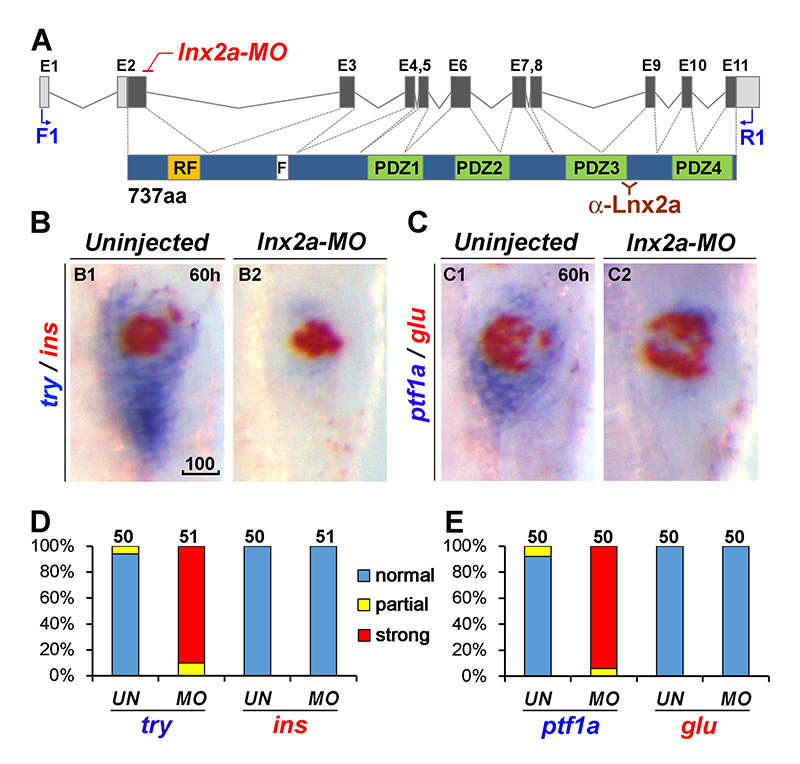
Click image to enlarge.
Figure 4. lnx2a knock-down causes defects in the exocrine pancreas.
(A) Schematic drawing of the lnx2a locus containing 11 exons and the Lnx2a protein (737 aa) containing a RING-finger domain (RF), the Numb-binding NPAF motif (F), and four PDZ domains. The translation initiation site is located in exon 2. The lnx2a-MO targets the splice donor site of exon 2. Primers in exon 1 (F1) and exon 11 (R1) are shown, as is the epitope for the Lnx2a antibody.
(B–C) The lnx2a-MO leads to inhibition of exocrine markers (try, ptf1a), but not endocrine markers (ins, glu) at 60 hpf.
(D–E) Quantification of marker expression. UN, uninjected; MO, lnx2a-MO; scale bar, 100μm.
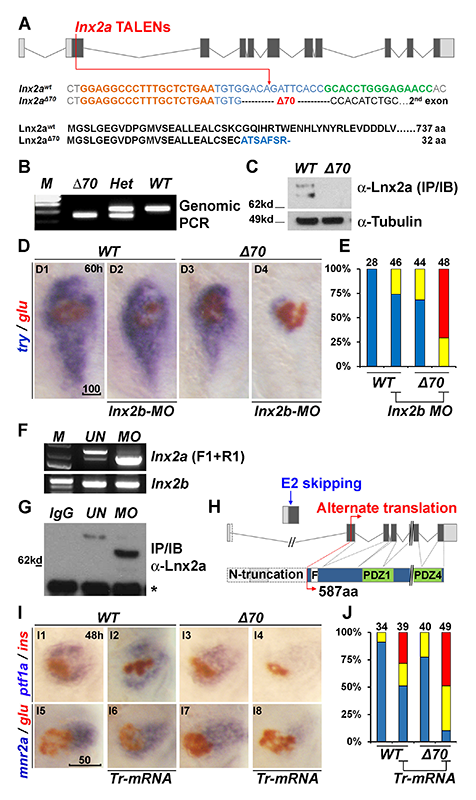
Click image to enlarge.
Figure 5. Generation of lnx2aΔ70 mutant using TALENs, and the functional redundancy of lnx2 genes
(A) Schematic representation of the lnx2a locus and the TALEN target site in exon 2. TALEN targets (left, orange and right, green), and the lnx2aΔ70 mutation are shown. Next, the protein sequences of wild-type and the lnx2aΔ70 frameshift allele are shown. Genotype of the lnx2aΔ70 mutant was analyzed by genomic PCR (B) and immuno-blotting of Lnx2a protein (C). (D) Marker gene (try and glu) expression shows little effect in lnx2aΔ70-null mutants or in lnx2b-MO–injected embryos. However, lnx2b-MO injection into lnx2aΔ70-mutant embryos shows suppression of exocrine markers. (E) Quantification of defects. (F–H) lnx2a-MO leads to production of N-truncated Lnx2a protein. (F) RT-PCR using primers F1 and R1 (Figure 4A) followed by sequencing shows that lnx2a-MO injection results in exon 2 skipping; lnx2b expression was unchanged. (G) Endogenous Lnx2a showed a smaller protein in lnx2a-MO–injected embryos. (H) Schematic drawing of exon 2 skipping, alternate translation start site, and N-truncated protein in lnx2a-MO–injected embryos. (I) N-truncated Lnx2a has an interfering effect; Tr-mRNA was injected into the WT and lnx2aΔ70–mutant embryos. (J) Quantification of pancreatic defects in (I). Scale bars: 100 mm (D1) and 50 mm (I5).
The most widely studied role of the ubiquitin pathway and E3 ubiquitin ligases is in the regulation of protein turnover. Numb is known as a target of mammalian Lnx proteins, and we confirmed that zebrafish Lnx2a can mediate the ubiquitination and degradation of Numb. Numb is a known inhibitor of the Notch signaling pathway, and Notch is an important factor in early pancreas formation. We thus hypothesized that depletion of Lnx2a and Lnxb leads to stabilization of Numb when it normally should be turned over, resulting in inhibition of Notch and thereby in a defect in pancreas cell differentiation. This view is strongly supported by the fact that a Numb-MO can rescue the loss of endocrine marker expression in lnx2a-splice MO–injected embryos and in lnx2adelta329–mutant fish (Figure 7).
Previous work by several laboratories may be summarized in our context as follows: (1) Notch signaling is required in pancreatic cell differentiation; (2) high Notch activity maintains precursor pools, while lower activity allows differentiation, in certain cases preceded by proliferation; (3) cells that downregulate Notch cannot maintain their precursor status. Our results led us to the following model for the role of Lnx2 proteins in pancreas development in zebrafish. We suggest that Lnx2a, together with Lnx2b, destabilizes Numb in ventral bud–derived cells, allowing Notch activity, which may be required for the specification and expansion of precursor cells. Loss of Lnx2 activity in the lnx2adelta329 mutant or lnx2a morphant stabilizes Numb to inhibit Notch in cells where it is normally active, ultimately interfering with the normal developmental progression of these cells. We suggest that, in normal development, the regulation of Numb protein stability by Lnx2a/b is an important component of the system that controls the levels of Notch activity during pancreas formation and the assignment of cells to different pancreatic lineages (Reference 4).
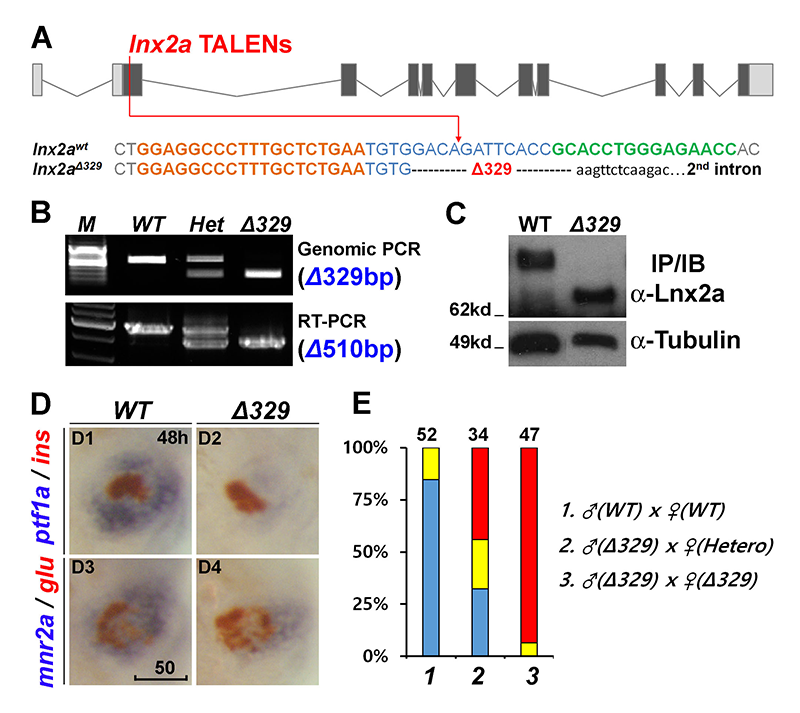
Click image to enlarge.
Figure 6. The lnx2aΔ329 mutant recapitulates the morphant phenotype.
(A) Sequence of the lnx2aΔ329 mutation. The mutant has lost the exon 2 splice donor site.
(B–C) Genotyping of lnx2aΔ329 by genomic PCR (Δ329), RT-PCR (Δ510 equaling exon 2) (B), and by Western blotting showing N-truncated protein (C).
(D) Phenotypic analysis of lnx2aΔ329 mutants at 48 hpf. (E) Quantification of D. Scale bar, 50mm.
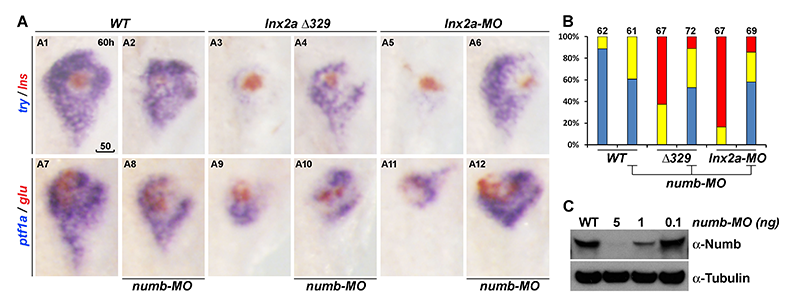
Click image to enlarge.
Figure 7. Ventral pancreas defects in lnx2aΔ329–mutant and lnx2a-MO–injected embryos can be rescued by knock-down of Numb.
(A) The translation-blocking MO for Numb was injected into WT, lnx2aΔ329–mutant, and lnx2a-MO–injected embryos, and pancreatic phenotype was examined at 60 hpf.
(B) Quantification of pancreatic defects by analysis of ptf1a expression. Exocrine pancreas defects were substantially rescued by knock-down of Numb; numb-MO had little effect in WT embryos.
(C) Immuno-blotting of embryo extracts with Numb antibody shows the efficiency of the numb-MO. Scale bar, 50 mm.
Publications
- Zarelli VE, Dawid IB. Inhibition of neural crest formation by Kctd15 involves regulation of transcription factor AP-2. Proc Natl Acad Sci USA 2013 110:2870-2875.
- Toyama R, Kim MH, Rebbert ML, Gonzales J, Burgess H, Dawid IB. Habenular commissure formation in zebrafish is regulated by the pineal gland-specific gene unc119c. Dev Dyn 2013 242:1033–1042.
- Kim MH, Rebbert ML, Ro H, Won M, Dawid IB. Cell adhesion in zebrafish embryos is modulated by March 8. PLoS One 2014 9:e94873.
- Won M, Ro H, Dawid IB. Lnx2 ubiquitin ligase is essential for exocrine cell differentiation in the early zebrafish pancreas. Proc Natl Acad Sci USA 2015 112(40):12426-12431.
- Wong TC, Rebbert M, Wang C, Chen X, Heffer A, Zarelli VE, Dawid IB, Zhao H. Genes regulated by potassium channel tetramerization domain containing 15 (Kctd15) in the developing neural crest. Int J Dev Biol 2016 60:159-166.
Collaborators
- Harold Burgess, PhD, Section on Behavioral Neurogenetics, NICHD, Bethesda, MD
- Raymond Habas, PhD, Temple University, Philadelphia, PA
- Hui Zhao, PhD, The Chinese University, Hong Kong, China
Contact
For more information, email dawidi@mail.nih.gov or visit http://sdb.nichd.nih.gov.


