Transcriptional and Translational Regulatory Mechanisms in Nutrient Control of Gene Expression

- Alan G. Hinnebusch, PhD, Head, Section on Nutrient Control of Gene Expression
- Hongfang Qiu, PhD, Staff Scientist
- Neelam Sen, PhD, Research Fellow
- Abdollah Ghobakhlou, PhD, Postdoctoral Fellow
- Suna Gulay, PhD, Postdoctoral Fellow
- Neha Gupta, PhD, Postdoctoral Fellow
- Anil Thakur, PhD, Postdoctoral Fellow
- Vishalini Valabhoju, PhD, Postdoctoral Fellow
- Fnu Yashpal, PhD, Postdoctoral Fellow
- Quira Zeidan, PhD, Postdoctoral Fellow
- Jinsheng Dong, PhD, Senior Research Assistant
- Fan Zhang, MS, Senior Research Assistant
- Laura Marler, BS, Graduate Student
We study the fundamental mechanisms involved in the assembly and function of translation initiation complexes for protein synthesis, using yeast as a model system to exploit its powerful combination of genetics and biochemistry. The translation initiation pathway produces an 80S ribosome bound to mRNA with methionyl initiator tRNA (tRNAi) base-paired to the AUG start codon. The tRNAi is recruited to the small (40S) subunit in a ternary complex (TC) with GTP–bound eIF2 to produce the 43S preinitiation complex (PIC) in a reaction stimulated by eIFs 1, 1A, 3, and 5. The 43S PIC attaches to the 5′ end of mRNA, facilitated by cap-binding complex eIF4F (comprising eIF4E, eIF4G, and the RNA helicase eIF4A) and poly(A)-binding protein (PABP) bound to the poly(A) tail, and scans the 5′ untranslated region (UTR) for the AUG start codon. Scanning is promoted by eIFs 1 and 1A, which induce an open conformation of the 40S and rapid TC binding in a conformation suitable for scanning successive triplets entering the ribosomal P site (P-out), and by eIF4F and other RNA helicases, such as Ded1, that remove secondary structure in the 5′ UTR. AUG recognition evokes tighter binding of the TC in the P-in state and irreversible GTP hydrolysis by eIF2, dependent on the GTPase–activating protein (GAP) eIF5, releasing eIF2-GDP from the PIC with tRNAi remaining in the P site. Joining of the 60S subunit produces the 80S initiation complex ready for protein synthesis. Our current aims in this research area are to (1) elucidate functions of eIF1, eIF1A, eIF2, and 40S proteins in TC recruitment and start codon recognition; (2) identify distinct functions of RNA helicases eIF4A (and its cofactors eIF4G/eIF4B), Ded1, and Dbp1 in mRNA activation, 48S PIC assembly, and scanning in vivo; (3) uncover the mechanisms of translational repression by the repressors Scd6, Pat1, Dhh1, and Khd1; (4) elucidate possible functions of yeast orthologs of eIF2A and eIF2D in eIF2–independent initiation of translation in stress conditions; (5) elucidate the in vivo functions of Rli1/ABCE1 (translation initiation factor/ATP-binding cassette E1, a ribonuclease inhibitor) and of yeast orthologs of eIF2D and the MCT-1/DENR complex (a translational enhancer) in ribosome recycling at stop codons in vivo.
We also analyze the regulation of amino acid–biosynthetic genes in budding yeast as a means of dissecting fundamental mechanisms of transcriptional control of gene expression. Transcription of these genes is coordinately induced by the activator Gcn4 during amino acid limitation owing to induction of Gcn4 at the translational level. The eviction of nucleosomes that occlude promoter DNA sequences and block access by RNA polymerase is thought to be a rate-limiting step for transcriptional activation. Previous studies implicated certain histone chaperones, ATP–dependent chromatin-remodeling complexes, or histone acetyltransferase complexes in eviction of promoter nucleosomes at certain yeast genes, but it was unclear whether these co-factors function at Gcn4 target genes. Our aim is to elucidate the full set of co-factors that participate in promoter nucleosome eviction at Gcn4 target genes, their involvement in this process genome-wide, and the transcriptional consequences of defective nucleosome eviction. Functional cooperation among the chromatin-remodeling complexes SWI/SNF, RSC, and Ino80 complex is the focus of current studies.
Structures of yeast preinitiation complexes reveal conformational changes from mRNA scanning to start-codon recognition.
It is thought that the scanning PIC assumes an open conformation and that AUG recognition evokes a closed state that arrests scanning with more stable tRNAi binding (P-in state), attendant displacement of the eIF1A-CTT (C-terminal tail) from the P site, and dissociation of eIF1 from the 40S subunit. In collaboration with Venki Ramakrishnan's and Jon Lorsch's groups, we obtained cryo-EM reconstructions of yeast PICs that represent different stages of the initiation pathway, at 3.5–6.1 Å resolution. These include 40S-eIF1–eIF1A complexes and partial yeast 48S PICs in open and closed conformations (py48S-open and py48-closed). The structures yield valuable information about conformational changes in the transition from scanning to AUG selection. The py48S-open complex, formed using mRNA with AUC start codon, reveals an upward shift of the 40S head that widens the mRNA entry channel and opens its latch, which should facilitate mRNA insertion into the binding cleft to form the scanning PIC. Moreover, the P site is widened and lacks tRNAi contacts with the 40S body present in canonical 80S-tRNAi complexes. Formed with mRNA(AUG), py48S-closed reveals downward head movement that closes the latch, clamps the mRNA into the binding cleft, and fully encloses tRNAi in the P site. The eIF1A N-terminal tail (NTT) interacts with the AUG:anticodon duplex, consistent with its role in stabilizing P-in. As a prelude to eIF1 dissociation from the 40S subunit, eIF1 is repositioned on the 40S and deformed to prevent a clash with tRNAi. Both py48S–open and –closed complexes reveal eIF2beta and portions of the eIF3 complex. The eIF3 trimeric subcomplex eIF3b-CTD/eIF3i/eIF3g-NTT resides on the subunit-interface surface of the 40S and appears to lock mRNA into the 40S binding cleft. Exclusively in py48S-open, eIF2beta interacts with tRNAi and segments of eIF1 and eIF1A, which should stabilize binding of TC and eIF1 to the scanning PIC prior to AUG recognition. Indeed, we showed that mutations at the eIF2beta:eIF1 interface increase aberrant recognition of UUG codons in vivo—the expected consequence of shifting the system from the open to closed conformations—thus establishing the physiological relevance of the py48S-open and py48S-closed structures. Recently, we showed that contacts between eIF1A-NTT residues and the AUG:anticodon duplex in the P site stabilize the closed conformation of the PIC, promoting stable tRNAi binding to reconstituted PICs formed with mRNA containing a near-cognate UUG start codon, and increasing utilization of poor initiation sites in vivo, including near-cognate start codons and AUG codons with poor surrounding sequence context. Some of these substitutions are recurrent mutations in human uveal melanomas. We also demonstrated that a clash between loop-2 of eIF1 and the D-loop of tRNAi, predicted to impede rearrangement of the PIC to the closed conformation without full accommodation of tRNAi in the P site at an AUG codon, is an important determinant of stringent selection of AUG triplets in favorable context as initiation codons.
eIF4B and DEAD-box RNA helicases Ded1 and eIF4A preferentially stimulate translation of long mRNAs with structured 5′ UTRs and low closed-loop potential but with weak dependence on eIF4G.
The RNA helicases eIF4A and Ded1 are believed to resolve mRNA secondary structures that impede ribosome attachment to the mRNA and scanning to the start codon, but whether they perform distinct functions in vivo was poorly understood. We compared the effects of mutations in Ded1 or eIF4A on translational efficiencies (TEs) by ribosome profiling. Despite similar reductions in bulk translation, inactivation of Ded1 substantially reduced the relative TEs of over 600 mRNAs, whereas inactivation of eIF4A similarly affected fewer than 40 mRNAs. Ded1–dependent mRNAs show greater than average 5′ UTR length and propensity for secondary structure, implicating Ded1 in scanning though structured 5′ UTRs. Thus, it appears that Ded1 is critically required for scanning through secondary structures in 5′ UTRs, while eIF4A promotes a step of initiation common to nearly all mRNAs, such as ribosome attachment. Ribosome profiling of a mutant lacking eIF4B showed that eliminating eIF4B preferentially impacts mRNAs with long structured 5′ UTRs and reduces the relative TEs of many more genes than does inactivation of eIF4A. The findings support an eIF4A–independent role for eIF4B in addition to its known function as eIF4A cofactor. Mutations in eIF4B, eIF4A, and Ded1 also preferentially impair translation of longer mRNAs in a manner mitigated by the ability to form closed-loop mRNPs via eIF4F-Pab1 association, suggesting cooperation between closed-loop assembly and eIF4B/helicase functions in stimulating initiation. Recently, we determined that the Ded1 paralog Dbp1 functionally cooperates with Ded1 in promoting translation of mRNAs with long, structure-prone 5′ UTRs in vivo. We identified genes in which the role of Ded1 is evident only in cells lacking Dbp1, and others in which Dbp1 function is unveiled by inactivation of Ded1. Moreover, Dbp1 overexpression can rescue translation of many Ded1–hyperdependent mRNAs. Interestingly, a small group of mRNAs with structurogenic sequences in coding regions are uniquely dependent on Dbp1.
Contacts of ribosomal uS3/Rps3 with mRNA at the 40S entry channel cooperate with PIC:mRNA interactions at the exit channel, requiring eIF3a to stabilize the closed conformation of the PIC at the start codon.
Collaborative work with the Lorsch laboratory, using the yeast reconstituted translation initiation system, identified distinct molecular functions for different domains/subunits of the 5-subunit eIF3 complex. Mutations throughout eIF3 were found to disrupt its interaction with the 43S PIC and diminish eIF3's ability to accelerate PIC recruitment to a native yeast mRNA. Alterations to the C-terminal domain (CTD) of the a-subunit and the eIF3b/i/g heterotrimeric module significantly slowed mRNA recruitment, and mutations within eIF3b/i/g destabilized binding of the eIF2.GTP.Met-tRNAi ternary complex to the PIC. Using model mRNAs lacking contacts with the 40S entry or exit channels, the eIF3a N-terminal domain was implicated in stabilizing PIC interactions with mRNA specifically at the 40S exit channel, while the eIF3a CTD plays an ancillary role at the entry channel. These functions are redundant, as defects at either channel can be rescued by filling the other channel with mRNA. The 40S ribosomal protein uS3 (yeast Rps3) makes direct contacts with the mRNA at the entry channel, and we showed that these contacts, involving two conserved arginine residues, are required for PIC–mRNA interactions at the entry channel. In the absence of these Rps3 arginines, PIC interactions with mRNA at the exit channel, mediated by the eIF3a–NTD, become essential for stable assembly of 43S-mRNA complexes in vitro. Substitutions of these Rps3 arginines also destabilize tRNAi binding to PICs assembled with mRNA harboring a UUG start codon in vitro, and they reduce initiation at UUG codons and a poor-context AUG codon in vivo. The results indicate that Rps3–mRNA interactions at the entry channel cooperate with mRNA interactions at the exit channel involving the eIF3a–NTD to promote 43S PIC binding to mRNA, and that PIC–mRNA interactions contribute to the overall stability of tRNAi binding to the P site of the closed conformation of the PIC and to the ability to utilize suboptimal initiation codons.
Click image to enlarge.
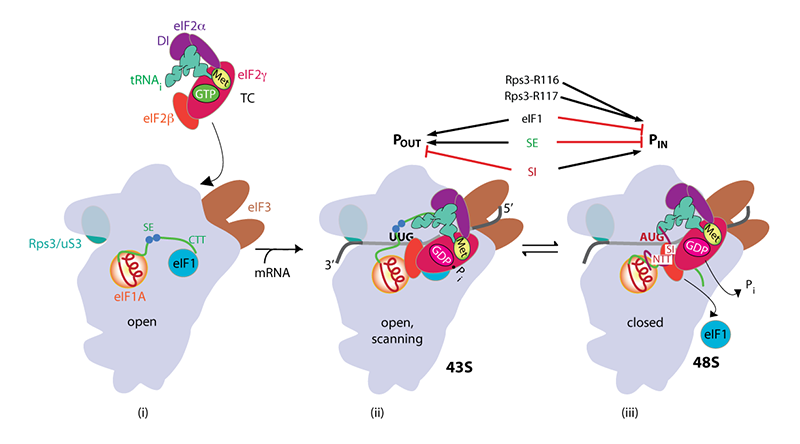
Rps3/uS3 plays a critical role in promoting mRNA binding at the 40S entry site and in stabilizing the preinitiation complex at the start codon.
A model describing known conformational rearrangements of the PIC during scanning and start codon recognition. (i) eIF1 and the scanning enhancers (SEs) in the C-terminal tail (CTT) of eIF1A stabilize an open conformation of the 40S subunit to which TC rapidly binds. Rps3 (uS3) is located on the solvent-exposed surface of the 40S near the entry channel; the bulk of eIF3 binds on the solvent-exposed surface with a prominent domain at the mRNA exit channel; (ii) The 43S PIC in the open conformation scans the mRNA for the start codon with Met-tRNAiMet bound in the POUT state. eIF2 can hydrolyze GTP to GDP•Pi, but release of Pi is blocked. (iii) On AUG recognition, Met-tRNAiMet moves from the POUT to the PIN state, clashing with eIF1 and the CTT of eIF1A, provoking displacement of the eIF1A CTT from the P site, dissociation of eIF1 from the 40S subunit, and Pi release from eIF2. The N-terminal tail (NTT) of eIF1A, harboring scanning inhibitor (SI) elements, adopts a defined conformation and interacts with the codon:anticodon helix. (Above) Arrows summarize that eIF1 and the eIF1A SE elements promote POUT and impede transition to PIN state, whereas the scanning inhibitor (SI) element in the NTT of eIF1A stabilizes the PIN state. (Below) In contact with mRNA at the entry channel, uS3/Rps3 residues R116/R117 stabilize the PIN state and also promote PIC interaction with mRNA at the entry channel, augmenting the role of eIF3 in PIC-mRNA interactions at the exit channel.
Interface between 40S exit channel protein uS7/Rps5 and eIF2α modulates start codon recognition in vivo.
Structures of yeast PICs have revealed that the β-hairpin of 40S ribosomal protein uS7/Rps5 is located in the 40S exit channel in proximity to mRNA nucleotides immediately 5′ of the start codon. Our previous analysis of substitutions in Rps5 had implicated Arg-225 and amino acids in the β-hairpin, particularly Glu-144, in stabilizing the closed conformation of the PIC in vitro, and revealed a requirement for these residues in enabling utilization of near-cognate (UUG) or AUG start codons in poor context in vivo. Comparison of yeast PICs in open or closed conformations further suggested that remodeling of the interface between Rps5 and domain 1 of eIF2α, one of the components of the eIF2-GTP-Met-tRNAi ternary complex, occurs in the transition from open to closed states. We showed that Rps5 substitutions disrupting eIF2α contacts, favored in the open complex, increase initiation at suboptimal initiation sites (UUG codons and poor-context AUG codons) in vivo, and that one such substitution, Rps5-S223D, stabilizes tRNAi binding to PICs reconstituted with mRNA harboring a UUG start codon in vitro, thus indicating inappropriate rearrangement to the closed conformation of the PIC at suboptimal start codons. Conversely, Rps5-D215 substitutions, perturbing Rps5-eIF2α interaction favored in the closed state, confer the opposite defects of suppressing utilization of poor initiation sites in vivo and (for D215L) of destabilizing tRNAi binding to reconstituted PICs. The results indicate that remodeling of the Rps5/eIF2α interface stabilizes first the open and then the closed conformation of the PIC to regulate the accuracy of start codon selection in vivo.
Genome-wide cooperation by histone acetyl transferase Gcn5, remodelers SWI/SNF and RSC, and chaperone Ydj1 in promoter nucleosome eviction and transcriptional activation
We investigated the mechanism of promoter nucleosome eviction during gene activation by analyzing the effects of mutations in one or more chromatin remodeling complexes, histone acetyltransferase (HAT) complexes, or histone chaperone on eviction of histone H3 at the large cohort of genes induced by transcription factor Gcn4 in response to amino acid starvation. By conventional chromatin immunoprecipitation analysis (ChIP) of four canonical Gcn4 target genes, ARG1, HIS4, ARG4, and CPA2, we excluded a requirement for several co-factors implicated previously at other genes (e.g., Asf1, Nap1, RSC) and implicated the remodeler SWI/SNF (Snf2), HAT Gcn5, and Hsp70 co-chaperone Ydj1 in nucleosome eviction at these Gcn4 target genes. Expanding our analysis genome-wide by H3 ChIP-Seq, we found that Snf2, Gcn5, and Ydj1 collaborate in evicting H3 from the –1 and +1 promoter nucleosomes and intervening nucleosome-depleted region (NDR) at a large fraction of the Gcn4 transcriptome; and that these three cofactors function similarly at the majority of yeast genes. Surprisingly, defective H3 eviction in co-factor mutants was coupled with reduced transcription (Pol II densities measured by Rpb3 ChIP-Seq) only for a subset of genes, which included the induced Gcn4 transcriptome and the most highly expressed constitutively expressed yeast genes. In fact, the most weakly expressed genes displayed higher relative transcription levels than other genes in response to global attenuation of nucleosome eviction in the mutants. The results established that steady-state eviction of promoter nucleosomes is required for maximal transcription of highly expressed genes and that Gcn5, Snf2, and Ydj1 function broadly in this step of gene activation, whereas some other aspect of transcriptional activation is more generally rate-limiting for transcription at the majority of genes in amino acid–deprived yeast. More recently, we uncovered extensive cooperation between SWI/SNF and a second chromatin remodeling complex, RSC, in evicting nucleosomes from different locations in the promoter, and in repositioning the –1 and +1 nucleosomes to produce wider NDRs, which are more highly depleted of nucleosomes, during transcriptional activation. We also found that SWI/SNF functions on par with RSC at the most highly transcribed subset of constitutively expressed genes, suggesting general cooperation by these remodelers in achieving maximum transcription rates in vivo.
Publications
- Qiu H, Chereji RV, Hu C, Cole HA, Rawal Y, Clark DJ, Hinnebusch AG. Genome-wide cooperation by HAT Gcn5, remodeler SWI/SNF, and chaperone Ydj1 in promoter nucleosome eviction and transcriptional activation. Genome Res 2016 26:211-225.
- Sen ND, Zhou F, Harris MS, Ingolia NT, Hinnebusch AG. eIF4B stimulates translation of long mRNAs with structured 5' UTRs and low closed-loop potential but weak dependence on eIF4G. Proc Natl Acad Sci USA 2016 113:10464-10472.
- Aitken CE, Beznosková P, Vlckova V, Chiu WL, Zhou F, Valášek LS, Hinnebusch AG, Lorsch JR. Eukaryotic translation initiation factor 3 plays distinct roles at the mRNA entry and exit channels of the ribosomal preinitiation complex. eLife 2016 5:e20934.
- Visweswaraiah J, Hinnebusch AG. Interface between 40S exit channel protein uS7/Rps5 and eIF2a modulates start codon recognition in vivo. eLife 2016 6:e22572.
- Dong J, Aitken CE, Thakur A, Shin BS, Lorsch JR, Hinnebusch AG. Rps3/uS3 promotes mRNA binding at the 40S ribosome entry channel and stabilizes preinitiation complexes at start codons. Proc Natl Acad Sci USA 2017 114:E2126-E2135.
- Hinnebusch AG. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem Sci 2017 42:589-611.
- Obayashi E, Luna RE, Nagata T, Martin-Marcos P, Hiraishi H, Singh CR, Erzberger JP, Zhang F, Arthanari H, Morris J, Pellarin R, Moore C, Harmon I, Papadopoulos E, Yoshida H, Nasr ML, Unzai S, Thompson B, Aube E, Hustak S, Stengel F, Dagraca E, Ananbandam A, Gao P, Urano T, Hinnebusch AG, Wagner G, Asano K. Molecular landscape of the ribosome pre-initiation complex during mRNA scanning: structural role for eIF3c and its control by eIF5. Cell Rep 2017 18:2651-2663.
Collaborators
- Katsura Asano, PhD, Kansas State University, Manhattan, KS
- David Clark, PhD, Section on Chromatin and Gene Expression, NICHD, Bethesda, MD
- Sergey E. Dmitriev, PhD, Lomonosov Moscow State University, Moscow, Russia
- Nicholas Guydosh, PhD, Section on mRNA Regulation and Translation, NIDDK, Bethesda, MD
- Nicholas Ingolia, PhD, University of California Berkeley, Berkeley, CA
- Jon Lorsch, PhD, Laboratory on the Mechanism and Regulation of Protein Synthesis, NICHD, Bethesda, MD
- Mercedes Tamame González, PhD, Instituto de Biología Funcional y Genómica, Universidad de Salamanca, Salamanca, Spain
- Venkatraman Ramakrishnan, PhD, MRC Laboratory of Molecular Biology, Cambridge, United Kingdom
- Gerhard Wagner, PhD, Harvard Medical School, Boston, MA
- Michael Zeschnigk, PhD, Institut für Humangenetik, Universitätsklinikum, Essen, Germany
Contact
For more information, email hinnebua@mail.nih.gov or visit http://sncge.nichd.nih.gov.


