Control of Ectodermal Development in Vertebrate Embryos

- Thomas D. Sargent, PhD, Head, Section on Vertebrate Development
- Neal MacDonald, AB, Postbaccalaureate Fellow
- Allisan Aquilina-Beck, BA, Technician (part time)
The laboratory focuses on mechanisms regulating the differentiation of cranial neural crest cells, which give rise to the bone and cartilage of the vertebrate jaw, neuro-cranium, and other structures of the face and head. Our approach is to manipulate transcriptional control mechanisms in the intact zebrafish embryo using gain- and loss-of-function strategies, with the aim of identifying the regulatory networks that control craniofacial development. Disruption of these developmental programs is the most common source of birth defects in humans, the detection, prevention, and treatment of which is a central aspect of NICHD’s mission. Equally important, understanding the regulation of gene expression in this complex embryonic tissue represents a challenging and fascinating problem in basic molecular and developmental biology.
The origin of the lab’s neural crest research was our discovery in 2003 that, in the frog Xenopus, the transcription factor TFAP2a is both necessary and sufficient to trigger the conversion of cells at the neural plate border from neural to neural-crest identity. We went on to show that TFAP2a mediates the transcriptional response to bone morphogenetic protein (BMP) signaling in neural-crest induction. We also carried out pioneering research on the homeodomain factor Dlx3, performing the first mouse knock-out of the gene encoding this factor and demonstrating the factor's function in the development of mammalian epidermis and placenta. We were also the first to demonstrate the phylogenetic conservation of Dlx gene–regulatory elements by transferring Xenopus Dlx enhancers into the mouse genome, showing that enhancers were conserved between these two distantly related vertebrates, including enhancers active in tissues, such as hair follicle and mammary gland, that have no counterpart in amphibians. We also investigated the role of Dlx3 and other members of this family in establishing the boundary of the neural crest in Xenopus. The Dlx family remains our current focus of research.
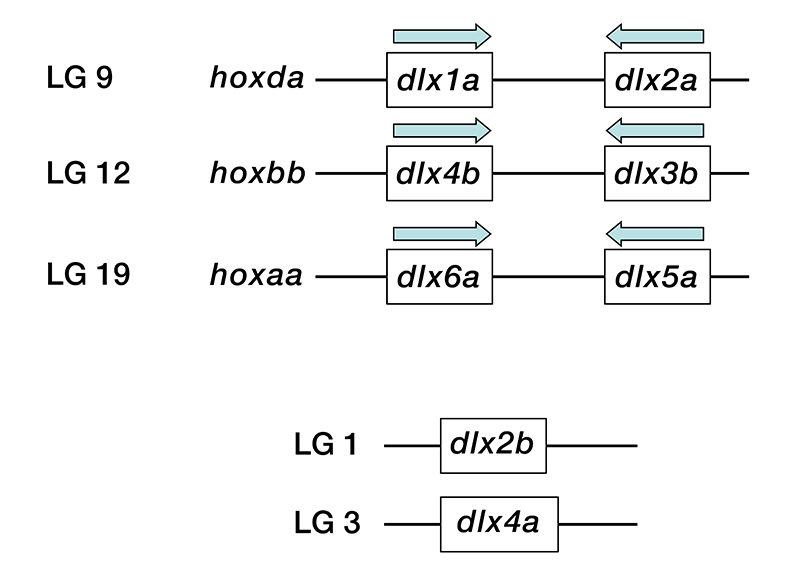
Click image to enlarge.
Figure 1. Genomic organization of dlx genes
Zebrafish dlx genes dlx1a, dlx2a, dlx3b, dlx4b, dlx5a, and dlx6a are organized into three pairs of convergently transcribed loci with about 10kb of intergenic 3′ sequence. Each pair is linked to one hox gene cluster, mirroring the pattern found in other vertebrates such as human. The remaining two, dlx2b and dlx4a, are located at unlinked positions in the genome. Drawing is not to scale.
Dlx gene function in cranial neural crest development
In the previous year we focused on loss-of-function experiments with the Dlx family of homeodomain transcription factors. Following reports from numerous labs, including our own, indicating that “knock down” analysis of gene function using antisense morpholinos frequently yielded results that were at variance with chromosomally based gene inactivation strategies, we set out to apply the CRISPR/Cas targeting approach to the dlx family in zebrafish. The objective of this project is to identify and characterize embryonic craniofacial phenotypes resulting from loss of individual dlx genes or combinations thereof. The targeting is being conducted in a line of fish carrying a GFP fluorescent marker expressed in cranial cartilage, enabling high-resolution imaging of affected tissues using confocal microscopy. So far, we identified INDEL (insertion and deletion) mutations that disrupt protein function of all six of the canonical dlx genes (i.e., dlx1–6), and we are in the process of targeting the two “extra” zebrafish Dlx loci (dlx2b and dlx4a). By in-crossing heterozygous carriers, we generated null embryos for all six individual dlx loci, as well as double-nulls for the linked pairs of genes (dlx1a+dlx2a, dlx3b+dlx4b, and dlx5a+dlx6a). In most cases, we observed little if any effect on early development. In particular, craniofacial cartilage formation was not affected. The exceptions were dlx2a and dlx1a/dlx2a double mutants, which showed abnormal jaw development in a portion of null embryos (i.e., incomplete penetrance). Most surprising is the observation that the dlx5a/dlx6a double-mutant embryos have apparently normal craniofacial development; equivalent mutations in the mouse result in massive defects in cranial morphology and limb formation. Our general conclusion is that, in zebrafish, dlx genes are much less critical for craniofacial or limb development (fins are also unaffected) than in the mouse. The finding has important evolutionary implications and is also relevant to the use of zebrafish as a general model system for functional analysis of genes found associated with disease in humans.
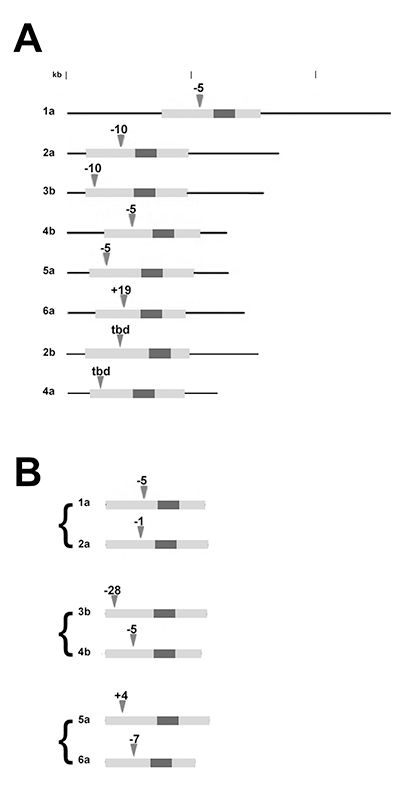
Click image to enlarge.
Figure 2. Summary of INDEL mutations in zebrafish dlx genes
A) The mRNAs of individual dlx genes are shown, with the open reading frames (ORFs) indicated in light gray and the homeodomains in dark gray. The location and size of deletions (-) or insertions (+) are indicated by the arrow heads. INDELs in dlx2b and dlx4a have not been fully characterized; tbd, to be determined.
B) Summary of the paired null mutations in linked loci (only ORFs are shown).
Publications
- Law SHW, Sargent TD. The serine-threonine protein kinase PAK4 is dispensable in zebrafish: identification of a morpholino-generated pseudophenotype. PLoS One 2014; E100268.
Collaborators
- Maria Morasso, PhD, Laboratory of Skin Biology, NIAMS, Bethesda, MD
Contact
For more information, email sargentt@mail.nih.gov or visit http://sargentlab.nichd.nih.gov.


