Mechanisms Regulating Interneuron Development

- Timothy J. Petros, PhD, Head, Unit on Cellular and Molecular Neurodevelopment
- Yajun Zhang, BM, Technician
- Maria Isaac, MS, Postbaccalaureate Fellow
The incredible diversity and heterogeneity of interneurons was observed over a century ago, with Ramon y Cajal hypothesizing in ‘Recollections of My Life’ that “The functional superiority of the human brain is intimately linked up with the prodigious abundance and unaccustomed wealth of the so-called neurons with short axons.” Although interneurons constitute the minority (20%) of neurons in the brain, they are the primary source of inhibition and are critical components in the modulation and refinement of the flow of information throughout the nervous system. Abnormal development and function of interneurons has been linked to the pathobiology of numerous brain diseases such as epilepsy, schizophrenia, and autism. Interneurons are an extremely heterogeneous cell population with distinct morphologies, connectivity, neurochemical markers, and electrophysiological properties, and, with the advent of new technologies to dissect gene expression and connectivity patterns, the classification of interneurons into specific subtypes is ever-evolving. However, our general understanding of the developmental mechanisms that generate this diversity remains largely poor. The goal of our lab is to dissect the genetic programs that determine the initial interneuron fate decisions during embryogenesis and to explore how the environment and genetic cascades interact to give rise to such stunning diversity of interneuron subtypes. We take a multifaceted approach to this issue, utilizing both in vitro and in vivo approaches to identify candidate mechanisms that regulate interneuron fate decisions. We strive to develop cutting edge techniques that will overcome the many challenges faced when studying interneuron development. Our ultimate goal is to discover the genetic cascades and signaling mechanisms that direct interneuron differentiation and maturation, which should act as a springboard for future research.
Click image to enlarge.
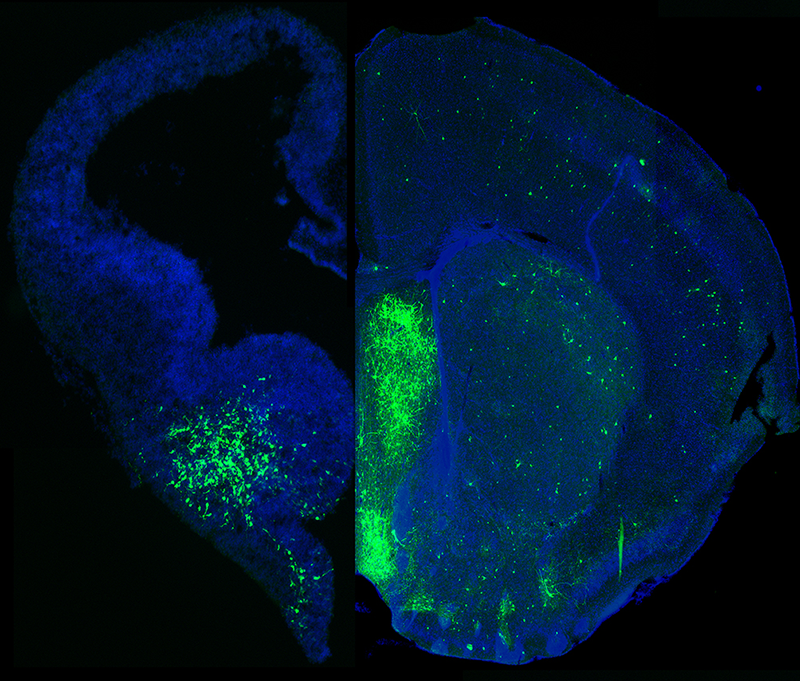
Figure 1. MGE–derived GABAergic cells populate many different brain regions.
Image depicts a section of an embryonic brain (left) that has been electroplated to label cells derived from the medial ganglionic eminence (MGE), merged with an section of an adult brain (right) displaying the incredible spatial and morphological diversity MGE–derived cells in the mature brain. Understanding how this heterogeneous population is generated from one embryonic brain structure is the focus of this laboratory.
Mechanisms regulating initial fate decisions within the medial ganglionic eminence
The medial ganglionic eminence (MGE) gives rise to the majority of forebrain interneurons, most notably the somatostatin- and parvalbumin-expressing (SST+ and PV+) subtypes. The MGE is a transient, dynamic structure that arises around E10 and bulges into the lateral ventricle over the next several days before dissipating at the end of embryogenesis. As initial fate decisions are generated within the MGE, there has been much focus on identifying a logic for interneuron generation from this region. Previous experiments characterized both a spatial and temporal gradient within the MGE that regulates the initial fate decision of becoming either PV+ or SST+ interneurons. SST+ interneurons are preferentially born early in embryogenesis from the dorso-posterior MGE whereas PV+ interneurons are born throughout embryogenesis with a bias of originating from the ventro-anterior MGE. Our work discovered an additional mechanism regulating this fate decision: the mode of neurogenesis. Using in utero electroporations, we found that PV+ interneurons are preferentially born from basal progenitors (also known as intermediate progenitors) whereas SST+ interneurons arise more commonly from apical progenitors. We hope build on this observation to discover how these distinct spatial, temporal, and neurogenic gradients coordinate to regulate initial fate decisions of MGE progenitors.
Click image to enlarge.
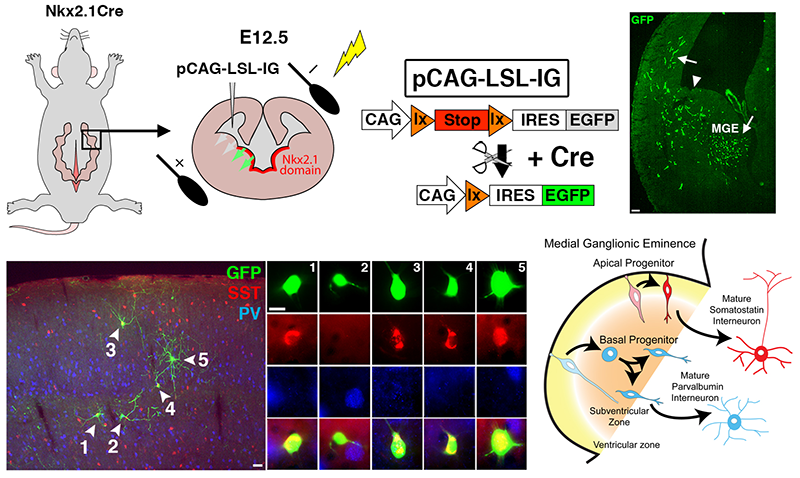
Figure 2. Manipulation of gene expression in the MGE by in utero electroporation (IUE)
Top. By using Nkx2.1-Cre mice, we can restrict expression of cre-dependent plasmids to the MGE. Note that the GFP+ cells are restricted to the MGE and cells migrating away from the MGE at 2 days post-electroporation.
Bottom. Representative example from a P25 cortex of a mouse whose MGE was electroplated at E12.5. In this series of experiments, we used IUE to demonstrate that the mode of neurogenesis plays a role in interneuron fate determination: apical neurogenic divisions are biased to generate somatostatin interneurons whereas basal divisions primarily give rise to parvalbumin cells.
How the environment sculpts interneuron diversity and maturation
Interneurons undergo an extensive tangential migration period before reaching their terminal brain region, upon which they interact with the local environment to differentiate and mature. The composition of interneuron subtypes varies significantly between different brain regions. Numerous experiments indicate that general interneuron classes, e.g., PV+– or SST+–expressing, are determined as cells become post-mitotic during embryogenesis, but when other features that define a mature interneuron subtype (neurochemical markers, cell type, and subcellular location of synaptic partners, electrophysiology properties, etc.) are established remains unknown. One hypothesis is that interneurons undergo an initial differentiation into ‘cardinal’ classes during embryogenesis, and maturation into ‘definitive’ subgroups requires active interaction with their mature environment. An alternate hypothesis is that immature interneurons are already genetically hard-wired into definitive subgroups, and that the environment more passively sculpts the maturation of these cells. To test these competing hypotheses, we are harvesting early postnatal interneuron precursors (P0-P2) in specific brain regions and transplanting them into wild-type hosts either homotopically (cortex-to-cortex) or heterotopically (cortex-to-hippocampus or cortex-to-striatum). The technique allows us to determine whether transplanted interneurons adopt properties of the host environment (indicating a strong role for the environment in regulating interneuron diversity) or retain subtype features more consistent with the donor region. Our initial experiments indicate that the environment largely determines the composition of interneuron subtypes in a brain region regardless of donor region. However, some interneuron subtypes appear to be more genetically predefined and resistant to environmental influences than others. In the future, we hope to combine this transplantation approach with other analytic techniques to more fully characterize how the environment sculpts interneuron diversity.
Click image to enlarge.
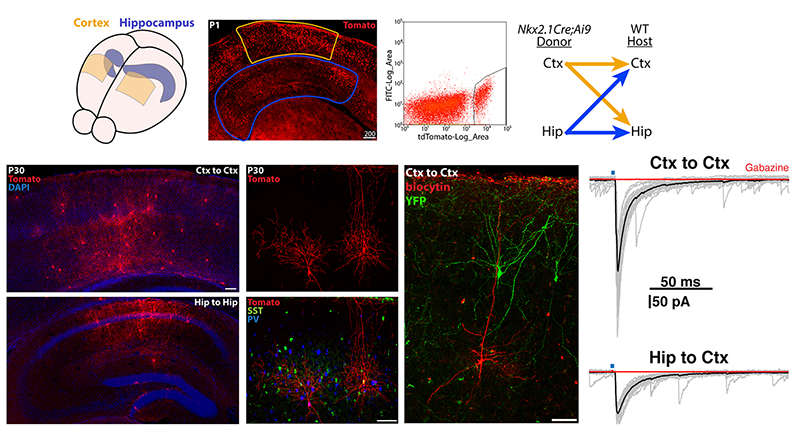
Figure 3. Transplantation of MGE–derived interneuron precursors into postnatal brains
Top. MGE–derived interneuron precursors are harvested from the cortex and hippocampus of P1 Nkx2.1-Cre;Ai9 mice, FACS–purified, and transplanted either homotopically (Ctx-to-Ctx, Hip-to-Hip) or heterotopically (Ctx-to-Hip, Hip-to-Ctx) into P1 wild-type (WT) mice.
Bottom. 30 days post-transplantation, tomato+ cells are dispersed throughout the host regions, displaying morphologies and neurochemical markers similar to endogenous interneurons. Grafted interneurons integrate into the host circuitry, as indicated by the postsynaptic responses in pyramidal cells upon stimulation of adjacent Nkx2.1-Cre;Ai32–derived, channel rhodopsin–expressing interneurons.
Novel approach to identify genetic cascades underlying interneuron fate decisions
The ability to longitudinally track gene expression within defined populations is essential for understanding how changes in expression mediate both development and plasticity. Previous screens that were designed to identify genes and transcription factors specific to SST– or PV–fated interneurons were largely unsuccessful because several issues significantly hinder these types of studies. First, these interneurons originate from the medial ganglionic eminence (MGE), which is a heterogeneous population of progenitors that give rise to both interneurons and a variety of GABAergic projection neurons, making it difficult to segregate interneuron progenitors from other cell types. Additionally, many markers that define mature interneuron subtypes are not expressed embryonically, and thus these class-defining markers are not helpful for studying MGE progenitors. In an ideal scenario, we would like to identify actively transcribed genes in MGE progenitors undergoing fate decisions while retaining the capacity to identify whether these cells become PV– or SST–expressing interneurons in the postnatal brain. To this end, we are developing a spatially and temporally inducible form of DNA adenine methylase identification (DamID) that will allow us to label the transcriptome of MGE progenitors. Labeled cells can be harvested at maturity once we have the tools to distinguish specific interneuron cell types. Then the methylated genomic DNA will be analyzed, allowing us to look back in time to identify candidate fate-determining genes expressed in specific interneuron populations. Our hope is that this strategy could be widely applicable so that an investigator could characterize the temporal gene expression pattern of the cell type of interest.
Click image to enlarge.
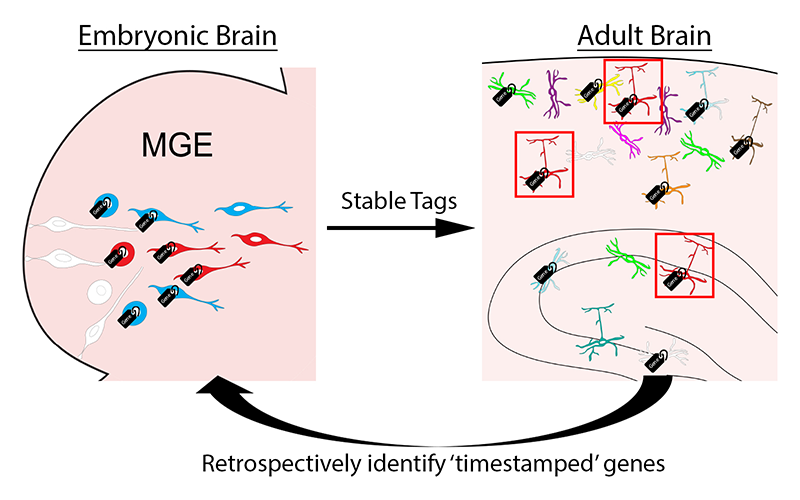
Figure 4. Timestamp of actively transcribed genes during development for future analysis
The goal of this approach is to label actively transcribed genes with stable methylation tags during embryogenesis as progenitors are undergoing initial fate decisions in the MGE. Then we can harvest specific interneuron subtypes in the adult brain using various transgenic mouse lines. Retrospective identification of actively transcribed gene during embryogenesis will provide us with candidate fate-determining genes for specific interneuron subtypes.
Contact
For more information, email tim.petros@nih.gov or visit https://science.nichd.nih.gov/confluence/display/petros.


