Development of the Vertebrate Circulatory System

- Brant M. Weinstein, PhD, Head, Section on Vertebrate Organogenesis
- Aniket Gore, PhD, Staff Scientist
- Van Pham, BS, Scientific Technician
- Marina Venero Galanternik, PhD, Postdoctoral Fellow
- Hyun Min Jung, PhD, Postdoctoral Fellow
- Mayumi Miller, PhD, Postdoctoral Fellow
- Scott Paulissen, PhD, Postdoctoral Fellow
- Laura Pillay, PhD, Postdoctoral Fellow
- Amber Stratman, PhD, Postdoctoral Fellow
- Natalie Aloi, BS, Postbaccalaureate Fellow
- Margaret Burns, BS, Postbaccalaureate Fellow
- Ciara Hu, BS, Postbaccalaureate Fellow
- Kelly Tomins, BS, Postbaccalaureate Fellow
The overall objective of this project is to understand how the elaborate networks of blood and lymphatic vessels arise during vertebrate embryogenesis. Blood vessels supply every tissue and organ with oxygen, nutrients, and cellular and humoral factors. Lymphatic vessels drain fluids and macromolecules from the interstitial spaces of tissues, returning them to the blood circulation, and they play an important role in immune responses. Our studies on the formation of blood and lymphatic vessels are of great clinical interest because of the roles both types of vessels play in cancer and ischemia. The zebrafish (Danio rerio) is a small tropical freshwater fish that possesses a unique combination of features that make it particularly suitable for studying vessel formation. Zebrafish are genetically tractable vertebrates with externally developing, optically clear embryos that are readily available for observation and experimental manipulation. Such features permit observation of every vessel in the living animal and simple, rapid screening for even subtle vascular-specific defects.
Our current studies use genetic screening, experimental analysis, and imaging to examine cues directing vascular patterning and morphogenesis, regulation of vascular integrity, assembly of the lymphatic system, and the roles of novel vascular-associated cells.
Tools for experimental analysis of vascular development in the zebrafish
The development of new tools to facilitate vascular studies in the zebrafish has been an important ongoing aim of this project. In previous work we (1) developed a widely used confocal micro-angiography method (Figure 1); (2) compiled an atlas of the anatomy of the developing zebrafish vasculature; (3) generated a variety of transgenic zebrafish lines expressing different fluorescent proteins within vascular or lymphatic endothelial cells, making it possible for us to visualize vessel formation in intact, living embryos; and (4) developed methodologies for long-term multiphoton confocal time-lapse imaging of vascular development in transgenic fish.
We are currently developing many new transgenic lines useful for in vivo vascular imaging as well as for in vivo blood or lymphatic endothelial-specific functional manipulation of signaling pathways involved in vascular specification, patterning, and morphogenesis. Notably, we generated “RiboTag” zebrafish, which facilitate high-throughput whole genome profiling of gene expression in blood and lymphatic vascular endothelium and in vascular smooth muscle cells, and we demonstrated that these transgenic lines can be used to perform in vivo profiling of vascular signaling pathways.
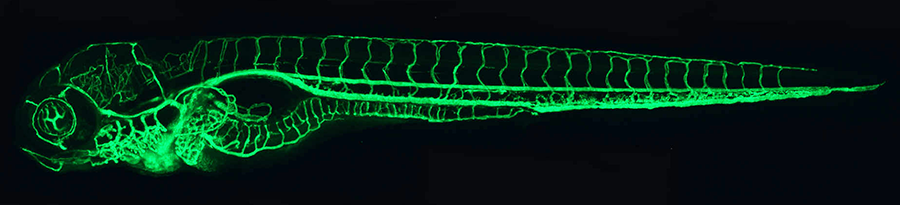
Click image to enlarge.
Figure 1. The zebrafish vascular system
Confocal micro-angiogram of the vascular system of a 4½-day-old zebrafish larva labeled by injecting fluorescent microspheres. The transparency of zebrafish larvae makes it possible to use high-resolution optical imaging methods to visualize the entire vasculature in exquisite detail.
Genetic analysis of vascular development
We use forward-genetic approaches to identify and characterize new zebrafish mutants that affect the formation of the developing vasculature. Using transgenic zebrafish expressing green fluorescent protein (GFP) in blood vessels (Figure 1) or lymphatic vessels, we are carrying out ongoing large-scale genetic screens for mutants induced by N-ethyl-N-nitrosourea (ENU). We have already identified hundreds of new vascular mutants with phenotypes that include loss of most vessels or subsets of vessels, increased sprouting/branching, and vessel mis-patterning. We recently carried out a new genetic screen to identify hemorrhagic stroke–susceptibility genes and used lymphatic-specific transgenic lines to perform screens for mutants specifically affecting the development of lymphatic vessels. We are pursuing the molecular cloning of the defective genes from all our new mutants, using next-generation whole-exome sequencing or RNAseq. To facilitate identification of the causative mutations (as opposed to naturally occurring polymorphisms) in our next-generation sequencing (NGS) data, we generated a large database of naturally occurring SNP (single nucleotide polymorphism) variants in three commonly used laboratory zebrafish strains. We also developed a web-based tool (“SNPfisher”) to facilitate querying and manipulating the database. Whole-exome sequencing combined with our SNP identification tools has already made it possibly to rapidly identify the causative mutations in some of our more recently identified mutants. The identification of additional defective genes from our mutants should result in further insights into the molecular mechanisms underlying vascular development and vascular integrity. Together, our ongoing mutant screens continue to yield a rich harvest of novel vascular mutants and genes, bringing to light new pathways critical for vascular development and vascular disease.
Analysis of vascular morphogenesis and integrity
Proper morphogenesis of vascular tubes and the maintenance of their integrity is of critical importance to human health. Malformation or rupture of vessels is the basis for stroke, the third leading cause of death and the most common cause of disability in developed nations. Intracerebral hemorrhage (ICH) accounts for 10 percent of stroke and is a particularly severe form of the disease, with disproportionately high rates of death and long-term disability. Therapeutic tools are still very limited, and prevention remains the most important way to reduce morbidity and mortality.
We used multiphoton time-lapse imaging to characterize patterns of vessel assembly throughout the developing zebrafish, and molecular and experimental analysis to understand how this pattern arises and what cues guide vascular specification, differentiation, and network assembly during development. Our discoveries included evidence that neuronal guidance factors play an important previously unknown role in vascular guidance and vascular patterning.
We also developed zebrafish models for ICH and uncovered a variety of different mutant zebrafish with defects in genes critical for vascular integrity (Figure 2).
Our current work includes projects aimed at (1) studying the specification, differentiation, and patterning of vascular smooth muscle in the zebrafish, making use of newly developed transgenic tools; (2) understanding the role of intracellular signaling substrates in regulating vascular endothelial signaling; (3) exploring the role of BMP–family ligands in modulating vessel growth and vascular integrity; and (4) analyzing additional pro- or anti-angiogenic factors.
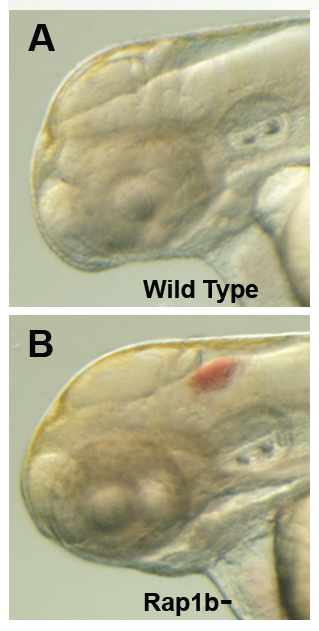
Click image to enlarge.
Figure 2. Intracranial hemorrhage (ICH) in the developing zebrafish
The clarity of zebrafish larvae also makes it straightforward to screen for animals with intracranial hemorrhage, as is evident in comparing lateral views of a 2-day-old wild-type larva (A) with a hemorrhage-prone larva deficient in rap1b (B).
Analysis of vascular patterning
We used multiphoton time-lapse imaging to characterize patterns of vessel assembly throughout the developing zebrafish. Our ongoing studies aim to understand how the patterns arise and what cues guide vascular network assembly during development. We previously demonstrated that known neuronal guidance factors play an important, previously unknown role in vascular guidance and vascular patterning, showing that semaphorin signaling is an essential determinant of trunk blood-vessel patterning (Figure 3). More recently, we also showed that chemokine signaling orchestrates the assembly and patterning of the developing lymphatic vasculature of the trunk. Current studies are elucidating the role of additional factors that guide the patterning of developing blood and lymphatic vascular networks in vivo, both in the trunk and in vascular beds in the eye, aortic arches, hindbrain, and other anatomical sites.
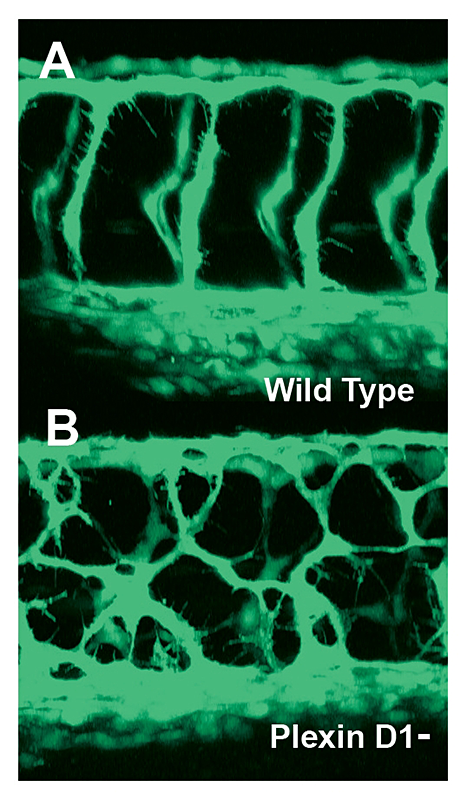
Click image to enlarge.
Figure 3. Mis-patterned trunk vessels in larvae lacking the vascular semaphorin receptor plexin D1
Confocal imaging of trunk vessels in a 2½-day-old wild-type (A) and a plexin D1–deficient (B) larva, showing loss of proper patterning of the trunk vessels caused by inability to receive semaphorin repulsive guidance signals.
Analysis of lymphatic development
The lymphatic system has become the subject of great interest in recent years because of the recognition of its important role in normal and pathological processes, but progress in understanding the origins and early development of the system has been hampered by difficulties in observing lymphatic cells in vivo and performing defined genetic and experimental manipulation of the lymphatic system in currently available model organisms. We showed that the zebrafish possesses a lymphatic system that shares many of the morphological, molecular, and functional characteristics of lymphatic vessels found in other vertebrates, providing a powerful model for the purpose of imaging and studying lymphatic development. As we continue to examine the origins and assembly of the lymphatic system of the zebrafish, we are developing new transgenic tools for imaging the development of the lymphatic system and for forward-genetic screening for lymphatic mutants. Our genetic analysis has already identified several novel genes involved in lymphatic development and patterning. We are also studying the roles of numerous genes required for specification, assembly, or patterning of the lymphatic endothelium, including a role for chemokine signaling in guidance and patterning of lymphatic vessel assembly in the developing larval trunk. We have also been studying a novel lymphatic-related perivascular cell population that plays a supportive role in the brain. Our ongoing studies will thus provide new insights into the molecular regulation of lymphatic development.
Epigenetic regulation of hematopoietic stem- and progenitor-cell emergence
We recently discovered a novel mechanism for epigenetic regulation of hematopoietic stem- and progenitor-cell (HSPC) specification. HSPCs emerge from the ventral wall of the dorsal aorta in all vertebrates. We found that a gene encoding DNA methyltransferase 3bb.1 (dnmt3bb.1) is expressed specifically in the ventral aortic endothelium, which gives rise to HSPCs. The gene functions downstream from a previously described genetic pathway for specification of HSPCs to promote the long-term maintenance of hematopoietic cell fate. Loss of the dnmt3bb.1 gene in vivo results in loss of HSPCs, while early ectopic overexpression of the dnmt gene is sufficient to induce ectopic hematopoietic gene expression. We are continuing to study the role of dnmt3bb.1 and DNA methylation in development and disease.
Publications
- Grainger S, Richter J, Palazón RE, Pouget C, Lonquich B, Wirth S, Grassme KS, Herzog W, Swift MR, Weinstein BM, Traver D, Willert, K. Wnt9a is required for the aortic amplification of nascent hematopoietic stem cells. Cell Rep 2016 6:1595-1606.
- Jung H-M, Castranova D, Swift MR, Pham VN, Galanternik MV, Isogai S, Butler MG, Mulligan TS, Weinstein BM. Development of the larval lymphatic system in the zebrafish. Development 2017 144:2070-2081.
- Krispin S, Melick CH, Stratman AN, Malinverno M, Stan RV, Gleklen J, Castranova D, Dejana E, Weinstein BM. Growth Differentiation Factor 6 (GDF6) promotes vascular stability by counteracting VEGF activity. Arterioscler Thromb Vasc Biol 2017 38(2):353-362.
- Stratman AN, Pezoa SA, Farrelly OM, Castranova D, Dye LE, Sidik H, Talbot WS, Weinstein BM. Mural-endothelial cell-cell interactions stabilize the developing zebrafish dorsal aorta. Development 2017 144:115-127.
- Venero Galanternik M, Castranova D, Gore AV, Jung HM, Stratman AN, Miller MF, Diemer J, Blewett N, Pham VN, Marquart G, Burgess H, Maraia R, Liu PP, Weinstein BM. A novel perivascular cell in the zebrafish brain. eLife 2017 6:e24369.
Collaborators
- Andreas Baxevanis, PhD, Computational and Statistical Genomics Branch, NHGRI, Bethesda, MD
- Harold Burgess, PhD, Section on Behavioral Neurogenetics, NICHD, Bethesda, MD
- George Davis, PhD, University of Missouri-Columbia, Columbia, MO
- Elisabetta Dejana, PhD, The FIRC Institute of Molecular Oncology Foundation, Milan, Italy
- Michael Granato, PhD, University of Pennsylvania, Philadelphia, PA
- Silvio Gutkind, PhD, Oral and Pharyngeal Cancer Branch, NIDCR, Bethesda, MD
- James Iben, PhD, Molecular Genomics Laboratory, NICHD, Bethesda, MD
- Sumio Isogai, PhD, Iwate Medical University, Morioka, Japan
- David Langenau, MD, Harvard Medical School, Boston, MA
- Paul Liu, MD, PhD, Genetics and Molecular Biology Branch, NHGRI, Bethesda, MD
- Richard Maraia, MD, Section on Molecular and Cell Biology, NICHD, Bethesda, MD
- Joan Marini, MD, PhD, Section on Heritable Disorders of Bone and Extracellular Matrix, NICHD, Bethesda, MD
- Yoh-suke Mukouyama, PhD, Laboratory of Stem Cell and Neuro-Vascular Biology, NHLBI, Bethesda, MD
- Xuetao Pei, MD, PhD, Beijing Institute of Transfusion Medicine, Beijing, China
- Lisa M. Price, PhD, Division of Developmental Biology, NICHD, Bethesda, MD
- David Raible, PhD, University of Washington, Seattle, WA
- Valya R. Russanova, PhD, Section on Chromatin and Gene Expression, NICHD, Bethesda, MD
- Radu V. Stan, MD, PhD, Geisel School of Medicine at Dartmouth, Lebanon, NH
- Zhaoxia Sun, PhD, Yale University School of Medicine, New Haven, CT
- William Talbot, PhD, Stanford University, Stanford, CA
- Jesús Torres-Vásquez, PhD, Skirball Institute of Biomolecular Medicine, New York, NY
- David Traver, PhD, University of California San Diego, La Jolla, CA
Contact
For more information, email weinsteb@mail.nih.gov or visit http://uvo.nichd.nih.gov.


