Modeling the Biophysics of the Membrane

- Alexander J. Sodt, PhD, Head, Unit on Membrane Chemical Physics
- Victor Chernomordik, PhD, Volunteer
- Kayla Sapp, PhD, Postdoctoral Fellow
- Christina M. Freeman, BS, Postbaccalaureate Fellow
- Mitchell Dorrell, BS, Predoctoral Fellow
- Julia Rakas, Summer Student
- Maya Wilson, Summer Student
The integrity of lipid membranes is essential for life. They provide spatial separation of the chemical contents of the cell and thus the enable the electrical and chemical potential differences that are used to transmit signals and perform work. However, the membrane must frequently be broken to form, for example, new membrane structures in the cell. The simplest structure is a vesicle to transport cargo. Such vesicles are constantly cycled between organelles and the outer plasma membrane. Thus, there is a careful balance between boundary-establishing membrane fidelity and the necessary ability of the cell to change these boundaries.
The challenge in studying the membrane is its complexity. The membrane is a thin sheet of small molecules, i.e., lipids. There are hundreds of types of lipids in the cell. Each lipid changes the properties of the membrane in its vicinity, sometimes making the sheet stiffer, sometimes softer, and sometimes acting to bend the membrane into a ball or tube. Furthermore, the lipids are constantly jostling and tangling with both each other and proteins embedded in the membrane. To predict of how membranes are reshaped thus requires not only knowing how lipids affect the properties of the membrane surface, but also the location of specific lipids.
A broad objective of our research is to create a publicly available software package that can be used either as a stand-alone application for analyzing membrane-reshaping processes or as a library for cellular-scale modeling packages for which the role of the membrane may be unclear or unanticipated. The model will incorporate the physical mechanisms we are investigating in the narrower objectives of the lab. These include: the influence of lipid curvature stress on the conformation of a protein; whether through-bilayer coupling can amplify a signaling state (for example, of a G protein–coupled receptor); and whether protein motifs (e.g., a thick hydrophobic region of a protein) can enhance the local lipid composition around a protein. These projects will address how the lipid micro-environment is controlled by the cell to change coupling between protein signaling states.
The projects use the NIH Biowulf computing cluster to run simulations and models. Molecular dynamics software (such as NAMD and CHARMM) are used to conduct molecular simulations. In-house software development for eventual public distribution is a key element of the lab's work.
Methodology to compare molecular simulations of bilayers to small-angle neutron scattering (SANS) experiments
We frame the question of “where are the lipids?” in terms of redistribution away from a null hypothesis in which lipids do not: (1) prefer to solvate a particular protein; (2) localize on structures of particular curvature (i.e., onto a vesicle); or (3) prefer to associate with lipids with specific chemistry. The broad lipid field literature has hypothesized extensions away from this null hypothesis based on fundamental biophysical experiments, namely that protein function varies based on local lipid competition; that lipid chemistry directly impacts the phase stability of vesicles and other curved phases; and that lipids can laterally “phase separate” based on preferred interactions between lipids. The challenge for research is that a lipid has a diameter less than a nanometer, while optical methods cannot characterize redistribution below approximately 200 nanometers.
This past year, we targeted small-angle neutron scattering (SANS) as a technique for determining the nano-scale co-localization of lipids. A SANS experiment averages all of the molecular correlations in a sample to a simple one dimensional intensity, essentially yielding an ensemble average of the Fourier transform of the internal structure. The effectiveness of the SANS experiment thus directly depends on the quality of the model used to explain the convoluted SANS intensity. We solved an important problem for modeling the SANS experiment: how to translate molecular simulations that employ periodic boundary conditions into the SANS intensity, while retaining lateral correlations. The method will be critical for interpreting the SANS intensity of sphingolipids; this includes a neutron spin echo experiment, which can show the timescale for bilayer fluctuations.
Modeling lipid redistribution governed by the peptide-lipid interface of gramicidin A
The question of how the lipid microenvironment changes protein function is a prime challenge in biophysics. A classic tool for this study is the small peptide gramicidin. Two gramicidin ‘monomers,’ which span a leaflet (half the bilayer), join to form a complete membrane-spanning channel. The channel is of a specific thickness that must be accommodated by the membrane. The accommodation deforms the membrane to match the channel. This simple two-state system thus responds exquisitely to a change in membrane properties, such as its thickness and stiffness. The most important lipids in this process are those directly next to the channel—they are perturbed the most, and thus the perturbation required is sensitive to their properties.
This project combined experiment, simulation, and theoretical analysis to describe how the lipid-peptide interface is differentially favored by lipids; in this case, it is favored by shorter lipids that match the peptide thickness. This was shown experimentally (by our collaborators in the Andersen lab) by comparing channel stability in two nearly equivalent bilayers: a bilayer with one lipid of medium height, and a mixture of short and tall lipids. Even though the average thickness of the two bilayers was equal, the channel stability was increased in the mixture. Theory and simulation not only co-validated the experimental finding, but provided molecular and material justification for the enhanced stability, also indicating how the channel would be stable vis-a-vis other changes in lipid chemistry, such as those leading to curvature preferences (Reference 3). In a third study (Reference 2), a connection was made between protein side-chain chemistry and the structure of the protein-lipid interface, namely, hydrogen bonding between peptide and lipid. Figure 1 is a cartoon showing the structure of the gramicidin channel, the molecular structure of the surrounding bilayer, and a depiction of the continuum model used to make predictions on a grander scale.
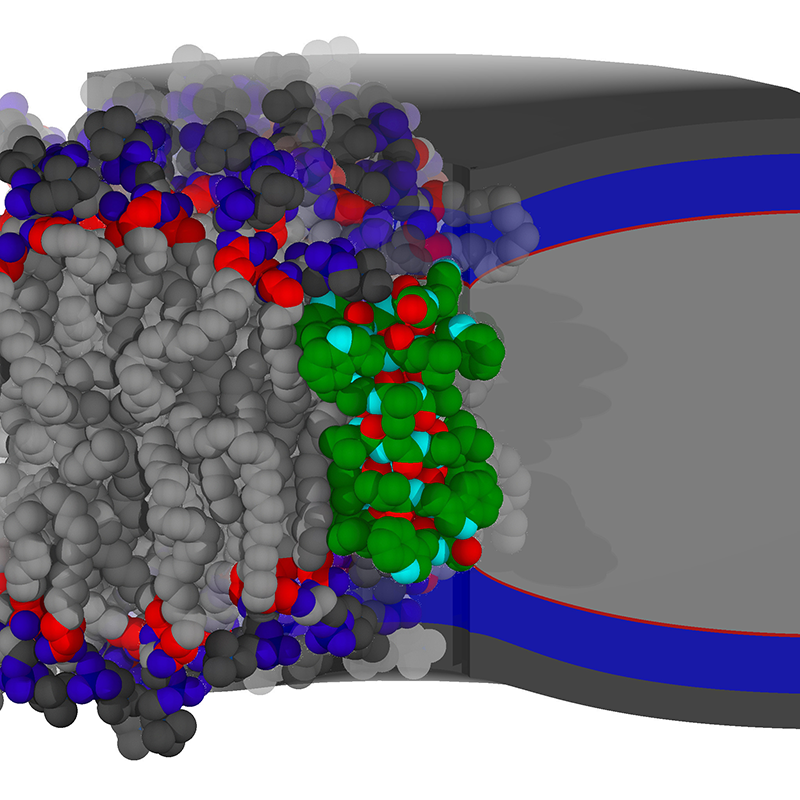
Click image to enlarge.
Figure 1. Continuum modeling of protein-membrane coupling
Shown in green is a short peptide channel deforming a thick bilayer in which it is embedded. At right is a continuum model of the bilayer color-coded by the layers of chemistry present. At left is a molecular simulation of the same peptide with equivalent coloring.
Publications
- Sodt A, Venable R, Lyman E, Pastor R. Nonadditive compositional curvature energetics of lipid bilayers. Phys Rev Lett 2016 117:138104.
- Beaven AH, Sodt AJ, Pastor RW, Koeppe RE, Andersen OS, Im W. Characterizing residue-bilayer interactions using gramicidin A as a scaffold and tryptophan substitutions as probes. J Chem Theory Comput 2017 13:5054-5064.
- Beaven AH, Maer AM, Sodt AJ, Rui H, Pastor RW, Andersen OS, Im W. Gramicidin A channel formation induces local lipid redistribution I: experiment and simulation. Biophys J 2017 112:1185-1197.
- Sodt AJ, Beaven AH, Andersen OS, Im W, Pastor RW. Gramicidin A channel formation induces local lipid redistribution II: a 3D continuum elastic model. Biophys J 2017 112:1198-1213.
- Chlanda P, Mekhedov E, Waters H, Sodt A, Schwartz C, Nair V, Blank PS, Zimmerberg J. Palmitoylation contributes to membrane curvature in Influenza A virus assembly and hemagglutinin-mediated membrane fusion. J Virol 2017 91:e00947-17.
- Gurnev PA, Roark TC, Petrache HI, Sodt AJ, Bezrukov SM. Cation-selective channel regulated by anions according to their Hofmeister ranking. Angew Chem Int Ed Engl 2017 129:3560-3563.
Collaborators
- Olaf Sparre Andersen, MD, Weill Medical College of Cornell University, New York, NY
- Sergey Bezrukov, PhD, Section on Molecular Transport, NICHD, Bethesda, MD
- Klaus Gawrisch, PhD, Laboratory of Membrane Biochemistry and Biophysics, NIAAA, Bethesda, MD
- Edward Lyman, PhD, University of Delaware, Newark, DE
- Joshua Zimmerberg, MD, PhD, Section on Integrative Biophysics, NICHD, Bethesda, MD
Contact
For more information, email alexander.sodt@nih.gov or visit http://sodtlab.nichd.nih.gov.


