Biophysics of Large Membrane Channels

- Sergey M. Bezrukov, PhD, DSci, Head, Section on Molecular Transport
- Tatiana K. Rostovtseva, PhD, Associate Scientist
- Philip A. Gurnev, PhD, Staff Scientist
- Maria Queralt-Martin, PhD, Visiting Fellow
- Amandine Rovini, PhD, Visiting Fellow
Healthy cell functioning and development require both precise compartmentalization and effective communication between cells and cell organelles. It is critically important for the systems to regulate metabolic transport, one of these being membrane channels. Deviant alterations in these complex dynamic structures often cause a shift from health to disease. Keeping this in mind, we study “large” beta-barrel channels of eukaryotes and bacteria, which are responsible for metabolite fluxes. With regard to their structure and mechanisms of selectivity and regulation, the channels are fundamentally different from the conventional ion channels studied by neurophysiology.
Specifically, we seek to understand the physical principles of their functioning under normal and pathological conditions. Among many wet-lab approaches, such as fluorescence correlation spectroscopy and confocal microscopy, our hallmark method is reconstitution of channel-forming proteins into planar lipid membranes that allows us to study them at the single-molecule level. Findings obtained in these experiments are rationalized within a framework of a physical theory of channel-facilitated transport that brings the knowledge necessary for intelligent drug design and other strategies to effectively correct aberrant interactions associated with disease.
To grasp the general principles of beta-barrel channel functioning and regulation, we work with a broad variety of proteins and peptides that form channels, including VDAC (Voltage-Dependent Anion Channel from the outer membrane of mitochondria), alpha-Hemolysin (toxin from Staphylococcus aureus), translocation pores of Bacillus anthracis (PA63), Clostridium botulinum (C2IIa) and Clostridium perfringens (Ib) binary toxins, Epsilon toxin (from Clostridium perfringens), OmpF (general bacterial porin from Escherichia coli), LamB (sugar-specific bacterial porin from Escherichia coli), OprF (porin from Pseudomonas aeruginosa), MspA (major outer-membrane porin from Mycobacterium smegmatis), Alamethicin (amphiphilic peptide toxin from Trichoderma viride), Syringomycin E (lipopeptide toxin from Pseudomonas syringae), and the bacterial peptide TisB involved in persister cell formation. We also use Gramicidin A (linear pentadecapeptide from Bacillus brevis) as a molecular sensor of membrane mechanical properties. With the goal of studying the channel-forming proteins under controlled conditions, we first isolate them from the host organisms, purify them, and then reconstitute them into planar lipid bilayers, the model membrane system with precisely defined physical properties. This allows us to observe channels interacting with the lipid membrane as modified by volatile anesthetics, by cytosolic proteins such as tubulin and alpha-synuclein, and by newly synthesized drugs that act as blockers of the translocation pores of bacterial toxins. Our motivation is that learning the molecular mechanisms of channel functioning is vital for the development of new approaches to treatment of various diseases, for which regulation of transport through ion channels plays the key role.
Structural features and lipid-binding domain of tubulin on biomimetic mitochondrial membranes
Tubulin has emerged as a highly unexpected component of mitochondrial membranes involved in regulation of membrane permeability through its inhibition of VDAC. The discovery of this functional role has reawakened interest in the nature of the tubulin-membrane interaction to answer a new question: how tubulin, a cytosolic protein known for its role in microtubule structure and dynamics, targets mitochondrial membranes. This year, using a combination of five biophysical methods—surface plasmon resonance, electrochemical impedance spectroscopy, bilayer overtone analysis, neutron reflectometry, and molecular dynamics simulations—we studied binding of tubulin to biomimetic membranes of different lipid compositions. We found that tubulin distinguishes between lamellar and nonlamellar lipids through a highly conserved amphipathic binding motif (Figure 1). Specifically, alpha-tubulin targets cell and organelle membranes by sensing lipid-packing defects, with broad consequences for both normal cellular function and disease. We showed that tubulin belongs to the group of peripherally bound amphitropic proteins, the subfamily of peripheral membrane proteins that interact directly with the lipid membrane rather than with intrinsic membrane proteins and are therefore strongly influenced by lipid composition. Several diseases, such as atherosclerosis, type II diabetes, and lysosomal storage disorders, are associated with defective distribution of intracellular lipids. Although involvement of amphitropic proteins in various cell functions has been unequivocally established, the mechanisms of their interaction with cellular membranes are only beginning to be understood because their weak reversible binding to the membranes creates obvious experimental difficulties in assessing binding conformations and kinetics.
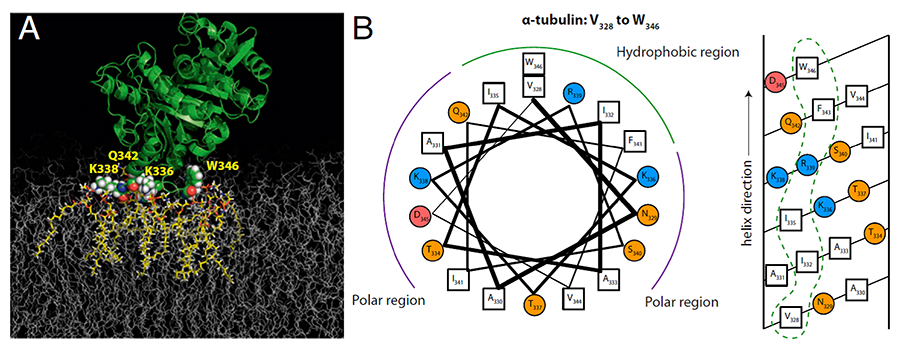
Click image to enlarge.
Figure 1. Tubulin binding residues identified by molecular dynamics (MD) simulations (Reference 1)
The proposed binding residues in alpha-tubulin (shown as van der Waals spheres) on the DOPE (dioleyl phosphatidylethanolamine) membrane (A). The binding residues were in direct contact with lipids (highlighted in yellow) for the entire duration of the simulation (about 1 ms). Helical wheel (left) and helical net (right) diagrams for the putative amphipathic binding region at the alpha end of the tubulin heterodimer (B). Hydrophobic residues are denoted by open squares, polar uncharged residues by orange circles, and positively and negatively charged residues by blue and red circles, respectively. Dashed loops indicate membrane-binding surfaces.
Partitioning of water-soluble polymers into beta-barrel channels in their functional states
Understanding polymer partitioning into nanoscale cavities of differing natures is important for many technological applications that include, but are not limited to, analytical chromatography, separation techniques, and purification methods. It is also critical in the qualitative interpretation and quantitative analysis of molecular interactions and biological regulation in the crowded cellular environment. This necessitates model studies with polymer solutions explored in both dilute and semidilute regimes. Recently, we studied polymer partitioning from semidilute water-based solutions of poly(ethylene glycol) (PEG) mixtures into a number of membrane-spanning beta-barrel channels of various origins, and the results were rationalized within the earlier formulated “polymers-pushing-polymers” model of nanopore partitioning. The model is based on the assumption that the larger component of the polymer mixture, being preferentially excluded from the channel nanopore, pushes the smaller component into the nanopore, thus representing forced polymer redistribution between the bulk and the channel. This year, we explored polymer mixtures by using two different methods, small-angle neutron scattering and nanopore conductance fluctuation analysis (Figure 2), to quantify the larger polymer parameters in the bulk and the degree of its partitioning in the pore, respectively. We first showed that the reduction of the PEG 3400 characteristic size with its increasing concentration in the bulk is statistically significant but small. We then demonstrated that partitioning of the larger polymer in the nanopore is negligible if its relative weight fraction is kept under 50%, in excellent agreement with the major assumptions of the model mentioned above. In our opinion, demonstration of this selective, “forced” partitioning of the small polymers into protein cavities to a significant excess of their bulk concentration is a step forward to a better qualitative and quantitative understanding of molecular crowding effects. Furthermore, it represents a new type of regulation of macromolecular transport, which may be of practical importance in the design and directed delivery of water-soluble drugs.
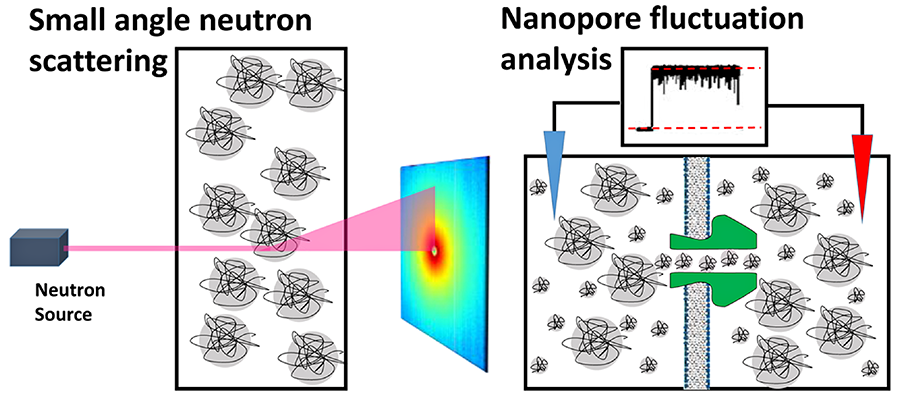
Click image to enlarge.
Figure 2. Cartoons of small-angle neutron scattering from bulk solutions (left) and nanopore conductance fluctuation analysis (right) (Reference 2)
These methods were used to probe structural and dynamic features of poly(ethylene glycol) in water/salt solutions in the dilute and semidilute regimes.
Ion specificity of the Hofmeister ranking
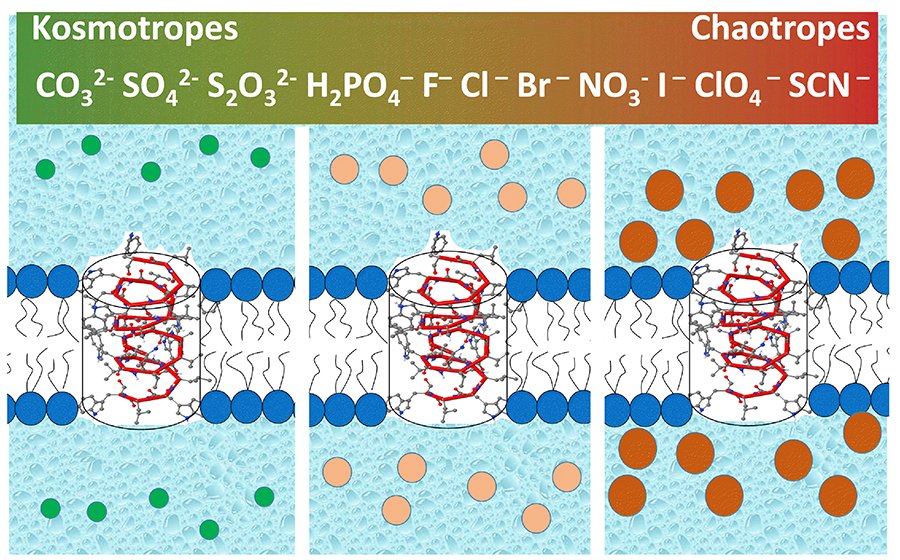
Ion specificity and, in particular, the distinctive effects of anions in salt-induced protein precipitation have been known since the 1880s, when Franz Hofmeister established the ranking of anions in their ability to precipitate egg yolk protein from water solutions. Experimental and theoretical studies have given a detailed empirical picture of the phenomenon, but the nature of the ionic interactions with the surfaces leading to the Hofmeister effect is still under debate. The only consensus is that it cannot be explained by standard theories of electrolytes. For example, bromide is unique in that its salts were recognized as a drug to treat epilepsy a couple of dozen years before Hofmeister’s studies and they are still in use to treat specific types of refractory seizures in children, but the mechanism of their action remains elusive. Although the Hofmeister ranking of salts has been a frequent target of biological studies including channel-facilitated membrane transport, this year we decided to take advantage of arguably the simplest ion transport model of modern biophysics—the channel formed by a linear pentadecapeptide, gramicidin A. Counter-intuitively, we found that conductance of this perfectly cation-selective channel increases about two-fold in the “Hofmeister anion series” H2PO4<Cl=Br=NO3<ClO4<SCN. Channel dissociation kinetics show even stronger dependence, with the dwell time increasing about 20-fold. While the conductance can be quantitatively explained by the changes in membrane surface potential owing to exclusion of kosmotropes from (or accumulation of chaotropes at) the surface (Figure 3), the kinetics proved to be more difficult to treat. We estimated the effects of changes in the energetics at the bilayer surfaces on the channel dwell time, concluding that the change would have to be greater than typically observed for the Hofmeister effect outside the context of the lipid bilayer. We believe that our results are of importance for further progress in understanding ion specificity, which manifests itself in many physicochemical and biological phenomena, including more than century-old medical applications.
Click image to enlarge.
Figure 3. Gramicidin A pore in a lipid membrane bathed by anions of different polarizability (Reference 3)
Both the conformational dynamics and conductance of this purely cation-selective peptide pore are regulated by anions in correlation with their position in the Hofmeister series by preferential depletion of kosmotropic anions (or accumulation of chaotropic anions) at the membrane-water interface.
Publications
- Hoogerheide DP, Noskov SY, Jacobs D, Bergdoll L, Silin V, Worcester D, Abramson J, Nanda H, Rostovtseva TK, Bezrukov SM. Structural features and lipid binding domain of tubulin on biomimetic mitochondrial membranes. Proc Natl Acad Sci USA 2017;114:E3622-E3631.
- Gurnev PA, Stanley CB, Aksoyoglu MA, Hong K, Parsegian VA, Bezrukov SM. Polyethylene glycols in semidilute regime: radius of gyration in the bulk and partitioning into a nanopore. Macromolecules 2017;50:2477-2483.
- Gurnev PA, Roark TC, Petrache HI, Sodt AJ, Bezrukov SM. Cation-selective channel is regulated by anions according to their Hofmeister ranking. Angew Chem Int Ed Engl 2017;56:3506-3509.
- Hoogerheide DP, Gurnev PA, Rostovtseva TK, Bezrukov SM. Mechanism of alpha-synuclein translocation through a VDAC nanopore revealed by energy landscape modeling of escape time distributions. Nanoscale 2017;9:183-192.
- Berezhkovskii AM, Dagdug L, Bezrukov SM. Mean direct-transit and looping times as functions of the potential shape. J Phys Chem B 2017;121:5455-5460.
Collaborators
- Vicente M. Aguilella, PhD, Universidad Jaume I, Castellón, Spain
- Alexander M. Berezhkovskii, PhD, Division of Computational Bioscience, CIT, NIH, Bethesda, MD
- Susan K. Buchanan, PhD, Laboratory of Molecular Biology, NIDDK, Bethesda, MD
- Leonid V. Chernomordik, PhD, Section on Membrane Biology, NICHD, Bethesda, MD
- Leonardo Dagdug, PhD, Universidad Autónoma Metropolitana-Iztapalapa, Mexico City, Mexico
- David Hoogerheide, PhD, National Institute of Standards and Technology, Gaithersburg, MD
- Jennifer C. Lee, PhD, Biochemistry and Biophysics Center, NHLBI, Bethesda
- Ekaterina M. Nestorovich, PhD, The Catholic University of America, Washington, DC
- Sergei Y. Noskov, PhD, University of Calgary, Calgary, Canada
- Adrian Parsegian, PhD, University of Massachusetts Amherst, Amherst, MA
- Olga Protchenko, PhD, Liver Diseases Branch, NIDDK, Bethesda, MD
- Dan Sackett, PhD, Section on Cell Biophysics, NICHD, Bethesda, MD
- Alexander Sodt, PhD, Unit on Membrane Chemical Physics, NICHD, Bethesda, MD
- Gerhard Wagner, PhD, Harvard Medical School, Cambridge, MA
- Michael Weinrich, MD, Office of the Director, NICHD, Bethesda, MD
- David L. Worcester, PhD, National Institute of Standards and Technology, Gaithersburg, MD
- Joshua Zimmerberg, MD, PhD, Section on Cellular and Membrane Biophysics, NICHD, Bethesda, MD
Contact
For more information, email bezrukov@helix.nih.gov or visit http://smt.nichd.nih.gov.


