Receptors and Actions of Peptide Hormones and Regulatory Proteins in Endocrine Mechanisms

- Maria L. Dufau, MD, PhD, Head, Section on Molecular Endocrinology
- Raghuveer Kavarthapu, PhD, Staff Fellow
- Muruganath Kumar Raju, PhD, Postdoctoral Fellow
- Peng Zhao, PhD, Postdoctoral Fellow
We investigate the molecular basis of peptide hormone control of gonadal function, with particular emphasis on the structure and regulation of the genes encoding the luteinizing hormone receptor (LHR) and prolactin (PRL) receptor (PRLR). We also investigate the regulatory mechanism(s) involved in the progression of spermatogenesis and the control of Leydig cell (LC) function. Our studies focus on the regulation of human LHR transcription (nuclear orphan receptors, epigenetics, DNA methylation, second messengers, repressors, corepressors, and coactivators), as well as on the multiple-promoter control of hPRLR gene transcription. We are elucidating the relevance of PRL, estradiol and its receptor (liganded or un-liganded), epidermal growth factor (EGF), the EGF receptors ERRBB1/EGFR and ERRB2/HER2 in the up-regulation of the PRLR, and their mechanistic commonalities for definition of PRL/PRLR–induced progression and metastasis of breast tumors and their role in persistent invasiveness in certain states refractory to adjuvant endocrine therapies. We also investigate novel gonadotropin-regulated genes relevant to the progression of testicular gametogenesis, LC function, and other endocrine processes. We focus on the function and regulation of the gonadotropin-regulated testicular RNA helicase (GRTH/DDX25), an essential post-transcriptional regulator of spermatogenesis, which was discovered, cloned, and characterized in our laboratory. The various functions of GRTH/DDX25 provide a fertile ground for the development of a male contraceptive.
The luteinizing hormone receptor
The luteinizing hormone receptor (LHR) is expressed primarily in the gonads, where it mediates LH signaling, which regulate cyclic ovarian changes and testicular function through its presence in the membrane of granulosa-luteal cells and Leydig cells, respectively. Human LHR gene transcription is controlled by complex and diverse networks, in which coordination and interactions between regulatory effectors are essential for silencing/activation of LHR expression. The proximal promoter site for the transcription factor Sp1 recruits histone (H) deacetylases (HDAC) and the Sin3A (a transcriptional regulatory protein) corepressor complex, which contribute to the silencing of LHR transcription. Site-specific acetylation/methylation–induced phosphatase release serves as an on switch for Sp1 phosphorylation at Ser641, which causes p107 repressor release from Sp1, recruitment of Transcription Factor II B (TFIIB) and RNA polymerase II (Pol II), and transcriptional activation. Maximal derepression of the gene is dependent on DNA demethylation of the promoter, on H3 acetylation, and on HDAC/Sin3A release. Positive Cofactor 4 (PC4) has an important role in the assembly of the preinitiation complex (PIC) in trichostatin A (TSA)–mediated LHR transcription. It is recruited by Sp1 following TSA treatment and acts as a coactivator. However, PC4 does not participate in TSA release of phosphatases, Sp1 phosphorylation, or release of repressor or complexes. Although TFIIB recruitment is dependent on PC4, we ruled out TFIIB as its direct target and acetylation of PC4 in the activation process. However, we demonstrated TSA–induced acetylation of PC4–interacting proteins, identified as acetylated H3 by mass spectrometry, and PC4's presence in the complex in association with chromatin at the promoter was demonstrated by ChIP/reChIP. Further MS/MS studies revealed association of PC4 with the H3.3 variant acetylated at several Lys residues. The presence of these modifications was further confirmed by site-specific H3 antibodies in western blots. ChiP/reChiP analysis showed an increased recruitment of complexes of PC4/acetylated H3 at these sites to the LHR promoter upon TSA stimulation. Immunoprecipitation studies of cells transfected with PC4-Flag by Flag antibody demonstrated interaction of PC4 with H3.3 induced by TSA, using H3.3–specific antibody, and the presence of the complex PC4-H3.3 at the LHR promoter was demonstrated by reChiP. Depletion of endogenous PC4 or H3.3 by siRNA caused marked reduction of TSA–induced formation of the complex, its recruitment to the promoter and of transcriptional activation of the LHR gene. Taken together, the findings indicate a critical role of PC4 association with acetylated H3.3 in TSA–induced Sp1–activated LHR transcription. The role of these interactions on chromatin structure and their participation in the assembly of the PIC and transcriptional activation are under investigation.
Gonadotropin-regulated testicular RNA helicase
Gonadotropin-regulated testicular RNA helicase (GRTH/DDX25) is a testis-specific member of the DEAD-box family of RNA helicases present in Leydig cells (LC) and meiotic germ cells and is essential for the completion of spermatogenesis. Males lacking GRTH are sterile owing to azoospermia resulting from failure of round spermatids to elongate. Besides its intrinsic RNA helicase activity, our studies demonstrated the essential participation of GRTH–mediated nuclear export/transport of mRNAs in the structural integrity of the Chromatoid Body (storage/processing of mRNAs), and of mRNAs transit/association to actively translating polyribosomes, where GRTH may regulate translational initiation of genes. GRTH is regulated by LH through the androgen (A)/androgen receptor (AR) at the transcriptional level in LCs (direct), with impact on hCG–induced steroidogenesis while not affecting basal circulating levels (of testosterone) in mice, and in germ cells (indirectly via AR in Sertoli cells); in germ cells GRTH's expression is both cell- and stage-specific. More recently, transgenic (tg) mouse models carrying GRTH 5′–flanking regions GFP provided in vivo systems that permitted differential elucidation of regions in the GRTH gene that direct its expression (upstream; –6.4/–3.6kb) in germ cells (pachytene spermatocytes and round spermatids) and downstream in LCs and GRTH's autocrine regulation by A/AR in LC and paracrine regulation in germ cells.
A functional binding site for the germ cell–specific transcription factor Germ Cell Nuclear Factor (GCNF), which is present in round spermatids (RS) and spermatocytes, resides in the distal region, –5270/–5252, of the GRTH gene, and we identified its paracrine regulation by A/AR, which is cell-specific and occurs exclusively in round spermatids (RS). The studies also provided evidence for the actions of androgen on GCNF cell-specific regulation of GRTH gene transcription/expression in RS. Moreover, GRTH associates with GCNF mRNA, and exerts of negative autocrine regulation of GCNF expression by reducing mRNA stability. Our in vivo and in vitro models link androgen's actions to germ cells through GCNF as an androgen-regulated transfactor that controls transcription of the GRTH gene. These important findings provided a connection of androgen action to two relevant germ-cell genes essential for the progress of spermatogenesis and established their regulatory inter-relationship (Reference 2).
Our early studies revealed that a missense mutation of R to H at amino acid 242 of GRTH found in 5.8% of patients with non-obstructive azoospermia, when express in COS1 cells, causes loss of the 61 KDa cytoplasmatic phospho-species with preservation of the nuclear 56 KDa non-phospho form. The finding provided an avenue to elucidate the function of phospho–GRTH in spermatogenesis. We generated a humanized mutant GRTH knock-in (KI) mouse. Very recent studies revealed that mutant KI mice are sterile with marked reduction in the size of the testes, which lack sperm, and with arrest at step 8 of round spermatids and complete loss of the phospho–GRTH species but with preservation of the non-phospho form. Furthermore, we demonstrated the requisite presence of the phospho–GRTH form to the integral functional structure of the chromatoid body. This mouse model will permit us to discern the biological and biochemical impact of the phospho-species in GRTH function and provide major novel insights for developing a male contraceptive.
The prolactin receptor
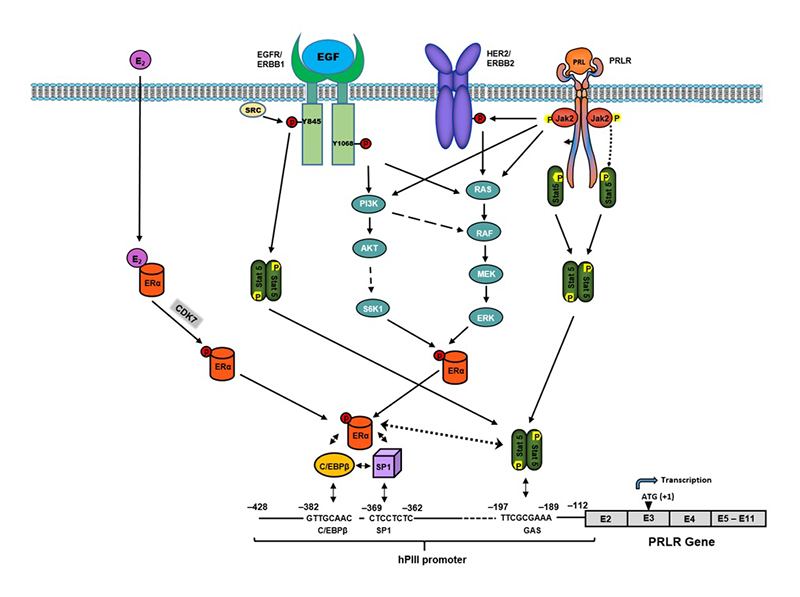
The human prolactin receptor (PRLR) mediates the diverse cellular actions of prolactin (PRL). PRL plays a major role in the proliferation and differentiation of breast epithelium and is essential for stimulation and maintenance of lactation. It plays an important role in the etiology and progression of breast cancer, tumoral growth, and chemo-resistance. PRLR expression is controlled at the transcriptional level by several promoters, one generic promoter (PIII), which lacks an estrogen response element (ERE) and is preferentially utilized, and five human-specific promoters (hPN1–hPN5), which we defined and characterized in our laboratory. Each promoter directs transcription/expression of a specific non-coding exon 1, a common non-coding exon 2, and coding exons (E3–E11). Complex formation of phosphorylated estrogen receptor alpha (ERa) homodimer (non-DNA bound) liganded or unliganded with the transcription factor Sp1 and dimers of the transcription factor C/EBPβ, bound to their sites at the PIII promoter, is required for basal (constitutive ERa homodimers) and estradiol (E2) or non-E2–induced transcriptional activation/expression of the human PRLR gene.
In tumoral breast, PRL causes cell proliferation by activating its cognate receptor. Exacerbation of PRL's actions in breast cancer, resulting from elevated receptor expression, can explain resistance to estrogen inhibitors in breast cancer. Using MCF7 breast cancer estrogen-responsive (ERa+)/oncogene HER2–positive (HER2+) cells, we demonstrated that, in the absence of E2, exogenous/endogenous PRL upregulated PRLR transcription/expression, with essential participation of ERa and of the JAK2/STAT5, mitogen-activated protein kinase (MAPK), and PI3K pathways. This occurs by interaction of phosphorylated ERa (generated by PRL/PRLR/JAK2), associated with Sp1 and C/EBPβ, with STAT5A and STAT5B bound to a nucleotide sequence, known as a GAS element, in the PIII promoter. We also found that ERRB/HER2, which is expressed in 10% of breast tumors, phosphorylated and activated by JAK2 via PRL/PRLR, induces ERa phosphorylation. Such cross-talk activation of ERBB2/HER2 signaling was identified as an alternate route for the PRLR increase, which is abolished by mutation of the GAS site (Stat5 DNA–recognition motif), by Stat5 siRNA, or by an ER antagonist (ICI). This indicates that ERa participates in PRLR transcription via PRL/PRLR/Stat5. PRL/PRLR induces phosphorylation of ERa through the JAK2/PI3K/MAPK/ERK– and HER2–activated pathways (Reference 1). Increased recruitment of phospho-ERa to Sp1 and C/EBPβ bound at promoter sites is essential for PRL–induced receptor transcription. Direct evidence for local actions of PRL independent of E2 is provided by the up-regulation of PRLR transcription/expression via the Stat5/ERa activation loop, with requisite participation of signaling mechanisms. These studies, which demonstrated a central role ERa in PRLR receptor up-regulation, are of relevance in states refractory to aromatase inhibitors, in which cancer progression can be fueled by endogenous PRL. Therapies that inhibit the function of PRL or PRLR, combined with inhibitors of various signaling pathways, could reverse resistance in breast cancer. Moreover, a combination therapy targeting ER and PRLR directly can offer an additional avenue to eliminate constitutive activation of ER and of PRLR by endogenous prolactin.
Paracrine inputs have an active role in breast tumor development, progression, and metastasis. Stromal fibroblasts secrete epidermal growth factor (EGF), which, through its receptor EGFR/ERRB1 present in breast tumors cells, activates signaling pathways, which in turn trigger requisite transcription factors and coactivators that can affect the proliferation of breast tumor cells. Other studies have addressed the role of EGFR in the up-regulation of the PRLR, given that most breast cancers that become resistant to endocrine therapy have elevated expression/activation of EGFR and its family member ERBB2. We showed, in MCF7 cells, marked activation of PRLR gene transcription/expression by exogenous EGF through EGFR1/ERBB1, independent of PRL/PRLR/JAK2 or E2, with essential involvement of the MAPK, ER1/2, and PI3K-AKT signaling pathways (Reference 3) for the recruitment of requisite phosphorylated transcription factors (ERa and STAT5b) to the PRLR PIII promoter. These pathways are mediated/activated by EGFR tyrosine kinase through p-tyrosines at 1068 and 1086 of the EGFR and by the tyrosine kinase cSRC–dependent phosphorylation of EGFR at tyrosine 845 for ERa and STAT5b phosphorylation, respectively. Apart from its independence of E2 and the activator requirements (PRL vs. EGF), there are important commonalities (prerequisite for ERa and STAT5) in the mechanism of PRLR transcription/expression (see below). Moreover, the studies revealed that STAT 5 interaction with ERa is essential for PRLR up-regulation. Our findings provide mechanistic avenues whereby, upon resistance to hormonal therapy, an increase of PRLR could promote progression and metastasis in breast cancer.
Further investigation revisiting our initial early findings on E2–induced upregulation of the PRLR, taking in to account our recent findings summarized above, have revealed the existence of at least three distinct up-regulatory modalities of PRLR in breast cancer cells relevant to breast cancer resistance and invasiveness. These relate to different inductors: (1) by endogenous PRL in the absence of E2; (2) by EGF in the absence of E2 and PRL; and (3) by E2 with PRL requirement. All these modalities of PRLR (1,2,3) transcription/expression, which participate in the upregulation of the PRLR, revealed commonalities. These are the central role of ERa phosphorylation via JAK2/PI3K/MAPK/ERK or JAK2/HER pathways induced by PRL/PRLR (1), or by EGF/ERBB1 signaling (2), or by CDK7 kinase induced by E2 (3). Another attribute they share is the requisite phosphorylation/activation of Stat5 (pStat5) by PRL/PRLR/JAK2 (1 & 3) and by EGF receptor tyrosine kinase induced by EGF (2). Also of relevance is the requisite interaction between pERa liganded or unliganded (associated with Sp1 and C/EBPb) and pStat5 bound to a GAS element at the PIII PRLR promoter. This interaction is required for productive stabilization of the ERa-Sp1-C/EBPb complex, which is essential for activation of PRLR promoter and transcription/expression of the PRLR receptor. With regard to modality 3, we demonstrated that the specific CDK7 kinase inhibitor (THZ1) significantly attenuates E2–induced phosphorylation at S118 and abrogates E2–induced PRLR transcription/expression. Moreover, the E2–induced cell migration was inhibited by PRL siRNA and THZ1, indicating its dependence on PRL/PRLR and CDK7, respectively. Our findings provide novel mechanistic insights into how the increase of PRLR in cancer maximizes the actions of endogenous/exogenous PRL that, upon resistance to hormonal therapy, could promote progression and metastasis in breast cancer (Figure 1). THZ1 treatment, singly or in combination with other inhibitory approaches targeting receptor function (PRLR, ERa, HER2, ERBB1) and/or the various signaling pathways (MAPK, PI3K, c-SRC, ERBB1 tyrosine kinase), with impact on ERa and Stat5 phosphorylation, could effectively ablate transcription/expression of PRLR fueled by tumor PRL and thus reverse or reduce resistance in breast cancer and, generally, its deleterious contribution to breast cancer.
Click image to enlarge.
Figure 1. Mechanisms of the up-regulation of hPRLR induced by E2/ERa, EGF/ERBB1, and PRL/PRLR.
Essential requirements for transcriptional activation of PRLR: phosphorylation of ERα (liganded or unliganded) and STAT5, which binds to its site (GAS) at the hPIII promoter and stabilizes the complex pERα/C/EBPβ/Sp1 by association with non-DNA–bound pERα, a requirement for recruitment of TFIIB and Pol II and PRLR transcriptional activation. Presented schematically are the modalities of PRLR up-regulation via E2/ERα with PRL/PRLR requirement: PRL/PRLR/JAK2/STAT5 per se; or through HER2 transactivation by JAK2 in the absence of E2. Also, EGF functional effects via its cognate receptor ERBB1, independent of PRL/PRLR, and E2 on the activation of intrinsic receptor tyrosine kinase and signal transduction pathways participating in STAT5 and ERα phosphorylation/activation, essential for PRLR gene transcription through hPIII promoter. Inhibition of ligand (E2)-dependent pERα (S118) by the specific covalent CDK7 inhibitor THZ1 causes abolition of ERα/C/EBPβ/SP1 complex formation at the promoter and of PRLR transcription/expression. The use of this inhibitor singly or in combination with other inhibitory approaches targeting receptor function (PRLR, ERα, HER2, ERBB1) and/or their signaling pathways could effectively ablate transcription of PRLR and its contribution to breast cancer (References 1,3,4).
Publications
- Kavarthapu R, Tsai Morris CH, Dufau ML. Prolactin induces up-regulation of its cognate receptor in breast cancer cells via transcriptional activation of its generic promoter by cross-talk between ERa and STAT5. Oncotarget 2014 5:9079-9091.
- Kavarthapu R, Dufau ML. Germ Cell Nuclear (GCNF/RTR) regulates transcription of gonadotropin-regulated testicular RNA helicase (GRTH/DDX25) in testicular germ cells—the androgen connection. Mol Endocrinol 2015 29:1792-1804.
- Kavarthapu R, Dufau ML. Role of EGF/ERBB1 in the transcriptional regulation of the prolactin receptor independent of estrogen and prolactin in breast cancer cells. Oncotarget 2016 7:65602-65613.
- Kavarthapu R, Dufau ML. Essential role of endogenous prolactin and cdk7 in estrogen-induced regulation of the prolactin receptor in breast cancer cells. Oncotarget 2017 8:27353-27363.
Collaborators
- James M. Pickel, PhD, Transgenic Core Facility, NIMH, Bethesda, MD
Contact
For more information, email dufau@helix.nih.gov or visit irp.nih.gov/pi/maria-dufau.


